Tune in to the latest #eLifePodcast to hear authors discuss the stories behind their recent discoveries https://t.co/awwqu5L3lE
eLife - the journal (@eLife)
Author Details
[Do not use this base pattern directly: see the following variants of this pattern for usage]
HTML
<div class="author-details" data-popup-contents="spuriousId" id="spuriousId">
<h4 class="author-details__name">[Do not use this base pattern directly: see the following variants of this pattern for usage]</h4>
</div>
Author Details Group
ICGC Chronic Myeloid Disorders Group
- Cancer Genome Project, Wellcome Trust Sanger Institute, Hinxton, United Kingdom
Competing interests
No competing interests declared- Luca Malcovati, Fondazione IRCCS Policlinico San Matteo, University of Pavia, Pavia, Italy
- Sudhir Tauro, Division of Medial Sciences, University of Dundee, Dundee, UK
- Jacqueline Boultwood, Nuffield Department of Clinical Laboratory Sciences, University of Oxford, UK
HTML
<div class="author-details" data-popup-contents="pageUniqueIdForThisAuthor1" id="pageUniqueIdForThisAuthor1">
<h4 class="author-details__name">ICGC Chronic Myeloid Disorders Group</h4>
<section class="author-details__section">
<ol class="author-details__list list list--bullet">
<li class="author-details__text">Cancer Genome Project, Wellcome Trust Sanger Institute, Hinxton, United Kingdom</li>
</ol>
</section>
<section class="author-details__section">
<h5 class="author-details__heading">Competing interests</h5>
<span class="author-details__text">No competing interests declared</span>
</section>
<ol class="list list--bullet">
<li>Luca Malcovati, Fondazione IRCCS Policlinico San Matteo, University of Pavia, Pavia, Italy</li>
<li>Sudhir Tauro, Division of Medial Sciences, University of Dundee, Dundee, UK</li>
<li>Jacqueline Boultwood, Nuffield Department of Clinical Laboratory Sciences, University of Oxford, UK</li>
</ol>
</div>
Author Details Person
Jenny Bloggs
- Evolutionary Studies Institute and Centre of Excellence in PalaeoSciences, University of the Witwatersrand, Johannesburg, South Africa
- School of Geosciences, University of the Witwatersrand, Johannesburg, South Africa
Present addresses
- Department of Inventive Inventions, Univertity of Wessex, Windowchester, Wessex
- Department of Underwater Basket Weaving, University of Somewhere
Contribution
Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article, Contributed unpublished essential data or reagentsContributed equally with
- Francesca Smith
- Wendel Jakes III
For correspondence
Competing interests
No competing interests declared
HTML
<div class="author-details" data-popup-contents="pageUniqueIdForThisAuthor1" id="pageUniqueIdForThisAuthor1">
<h4 class="author-details__name">Jenny Bloggs</h4>
<section class="author-details__section">
<ol class="author-details__list list list--bullet">
<li class="author-details__text">Evolutionary Studies Institute and Centre of Excellence in PalaeoSciences, University of the Witwatersrand, Johannesburg, South Africa</li>
<li class="author-details__text">School of Geosciences, University of the Witwatersrand, Johannesburg, South Africa</li>
</ol>
</section>
<section class="author-details__section">
<h5 class="author-details__heading">Present addresses</h5>
<ol class="author-details__list list list--bullet">
<li class="author-details__text">Department of Inventive Inventions, Univertity of Wessex, Windowchester, Wessex</li>
<li class="author-details__text">Department of Underwater Basket Weaving, University of Somewhere</li>
</ol>
</section>
<section class="author-details__section">
<h5 class="author-details__heading">Contribution</h5>
<span class="author-details__text">Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article, Contributed unpublished essential data or reagents</span>
</section>
<section class="author-details__section">
<h5 class="author-details__heading">Contributed equally with</h5>
<ol class="author-details__list list list--bullet">
<li class="author-details__text">Francesca Smith</li>
<li class="author-details__text">Wendel Jakes III</li>
</ol>
</section>
<section class="author-details__section">
<h5 class="author-details__heading">For correspondence</h5>
<ol class="author-details__list list list--bullet">
<li class="author-details__text"><a href="mailto:jenny@bloggs.com">jenny@bloggs.com</a></li>
<li class="author-details__text"><a href="tel:+1555432109876">+1 555-4321-09876</a></li>
</ol>
</section>
<section class="author-details__section">
<h5 class="author-details__heading">Competing interests</h5>
<span class="author-details__text">No competing interests declared</span>
</section>
<section class="author-details__section">
<span class="orcid">
<a href="https://orcid.org/0000-0002-3400-7927">
<picture>
<source srcset="../../assets/img/icons/orcid.svg" type="image/svg+xml">
<img src="../../assets/img/icons/orcid.png" class="orcid__icon"
alt="ORCID icon">
</picture> <span class="visuallyhidden">"This ORCID iD identifies the author of this article:"</span>
0000-0002-3400-7927</a>
</span>
</section>
</div>
Author Details Sub Group
ICGC Chronic Myeloid Disorders Group
- Cancer Genome Project, Wellcome Trust Sanger Institute, Hinxton, United Kingdom
Competing interests
No competing interests declaredSub-group 1
- Luca Malcovati, Fondazione IRCCS Policlinico San Matteo, University of Pavia, Pavia, Italy
- Sudhir Tauro, Division of Medial Sciences, University of Dundee, Dundee, UK
Sub-group 2
- Jacqueline Boultwood, Nuffield Department of Clinical Laboratory Sciences, University of Oxford, UK
HTML
<div class="author-details" data-popup-contents="pageUniqueIdForThisAuthor1" id="pageUniqueIdForThisAuthor1">
<h4 class="author-details__name">ICGC Chronic Myeloid Disorders Group</h4>
<section class="author-details__section">
<ol class="author-details__list list list--bullet">
<li class="author-details__text">Cancer Genome Project, Wellcome Trust Sanger Institute, Hinxton, United Kingdom</li>
</ol>
</section>
<section class="author-details__section">
<h5 class="author-details__heading">Competing interests</h5>
<span class="author-details__text">No competing interests declared</span>
</section>
<h5 class="author-details__secondary-name">Sub-group 1</h5>
<ol class="list list--bullet">
<li>Luca Malcovati, Fondazione IRCCS Policlinico San Matteo, University of Pavia, Pavia, Italy</li>
<li>Sudhir Tauro, Division of Medial Sciences, University of Dundee, Dundee, UK</li>
</ol>
<h5 class="author-details__secondary-name">Sub-group 2</h5>
<ol class="list list--bullet">
<li>Jacqueline Boultwood, Nuffield Department of Clinical Laboratory Sciences, University of Oxford, UK</li>
</ol>
</div>
Block Link
HTML
<div class="block-link">
<a href="#"
class="block-link__link">
<div class="block-link__inner">
Developmental Biology and Stem Cells
</div>
</a>
</div>
Block Link Archive Nav Link
HTML
<div class="block-link block-link--image">
<div class="block-link__picture_wrapper">
<picture class="block-link__picture">
<img src="https://unsplash.it/263/190"
alt=""
class="block-link__image"
>
</picture>
</div>
<a href="#"
class="block-link__link">
<div class="block-link__inner">
Developmental Biology and Stem Cells
</div>
</a>
</div>
Block Link Grid Listing
HTML
<div class="block-link block-link--grid-listing block-link--image">
<div class="block-link__picture_wrapper">
<picture class="block-link__picture">
<img src="https://unsplash.it/263/190"
alt=""
class="block-link__image"
>
</picture>
</div>
<a href="#"
class="block-link__link">
<div class="block-link__inner">
Developmental Biology and Stem Cells
</div>
</a>
</div>
Breadcrumb
- Research Article
HTML
<ul class="breadcrumbs">
<li class="breadcrumb-item">
<span class="breadcrumb-item__text">Research Article</span>
</li>
</ul>
Breadcrumb With Url
HTML
<ul class="breadcrumbs">
<li class="breadcrumb-item">
<a href="#" class="breadcrumb-item__link">Research Article</a>
</li>
<li class="breadcrumb-item">
<a href="#" class="breadcrumb-item__link">Magazine</a>
</li>
</ul>
Breadcrumb Without Url
- Research Article
- Magazine
HTML
<ul class="breadcrumbs">
<li class="breadcrumb-item">
<a href="#" class="breadcrumb-item__link">Research Article</a>
</li>
<li class="breadcrumb-item">
<span class="breadcrumb-item__text">Magazine</span>
</li>
</ul>
Button
HTML
<button class="button button--default" type="submit" id="button-id">Button</button>
Button Action Citation
HTML
<button class="button button--default button--action icon icon-citation" type="submit" id="button-action-citation" aria-label="Descriptive and accessible name">Button action (Citation)</button>
Button Action Comment With Hypothesis
HTML
<div data-hypothesis-trigger>
<button class="button button--default button--action icon icon-comment" type="submit" id="button-id" data-behaviour="HypothesisTrigger" aria-label="Descriptive and accessible name">Comment<span aria-hidden='true'><span data-visible-annotation-count></span> </span><span class='visuallyhidden'>Open annotations (there are currently <span data-hypothesis-annotation-count>0</span> annotations on this page). </span></button>
</div>
Button Action Comment
HTML
<button class="button button--default button--action icon icon-comment" type="submit" id="button-id" aria-label="Descriptive and accessible name">Button action (Comment)</button>
Button Action Download
HTML
<button class="button button--default button--action icon icon-download" type="submit" id="button-action-download" aria-label="Descriptive and accessible name">Button action (Download)</button>
Button Action Share
HTML
<button class="button button--default button--action icon icon-share" type="submit" id="button-action-share" aria-label="Descriptive and accessible name">Button action (Share)</button>
Button Action
HTML
<button class="button button--default button--action" type="submit" id="button-id" aria-label="Descriptive and accessible name">Button action</button>
Button Clipboard
HTML
<button class="button button--default" type="submit" id="button-id" data-behaviour="ButtonClipboard" data-clipboard="Text to store in clipboard" aria-label="Descriptive and accessible name">Button clipboard</button>
Button Extra Small
HTML
<button class="button button--default button--extra-small" type="button" id="button-id" name="button-name" aria-label="Descriptive and accessible name">Button extra small</button>
Button Full
HTML
<button class="button button--default button--full" type="button" id="button-id" name="button-name" aria-label="Descriptive and accessible name">Button full</button>
Button Inactive
HTML
<button class="button button--default button--inactive" type="button" id="button-id" name="button-name" aria-label="Descriptive and accessible name">Button inactive</button>
Button Link
HTML
<button class="button button--default" type="submit" id="button-id" aria-label="Descriptive and accessible name">Link button</button>
Button Login
HTML
<button class="button button--login" type="submit" id="button-id" aria-label="Descriptive and accessible name">Button extra small</button>
Button Outline
HTML
<button class="button button--outline" type="button" id="button-id" name="button-name" aria-label="Descriptive and accessible name">Button outline</button>
Button Reversed Extra Small
HTML
<button class="button button--reversed button--extra-small" type="button" id="button-id" name="button-name" aria-label="Descriptive and accessible name">Button reversed extra small</button>
Button Reversed Inactive
HTML
<button class="button button--reversed-inactive" type="button" id="button-id" name="button-name" aria-label="Descriptive and accessible name">Button reversed inactive</button>
Button Reversed Small
HTML
<button class="button button--reversed button--small" type="button" id="button-id" name="button-name" aria-label="Descriptive and accessible name">Button reversed small</button>
Button Reversed
HTML
<button class="button button--reversed" type="button" id="button-id" name="button-name" aria-label="Descriptive and accessible name">Button reversed</button>
Button Secondary
HTML
<button class="button button--default button--secondary" type="button" id="button-id" name="button-name" aria-label="Descriptive and accessible name">Button secondary</button>
Button Small
HTML
<button class="button button--default button--small" type="button" id="button-id" name="button-name" aria-label="Descriptive and accessible name">Button small</button>
Caption Text
[Do not use this base pattern directly: see the following variants of this pattern for usage]
HTML
<h6 class="caption-text__heading">[Do not use this base pattern directly: see the following variants of this pattern for usage]</h6>
Caption Text Additional Asset
Table 1 - source data 1.
Is the species name mentioned in the title, impact statement, abstract and digest of eLife papers (July–Sept 2014)?
HTML
<h6 class="caption-text__heading">Table 1 - source data 1.</h6>
<p class="caption-text__heading">Is the species name mentioned in the title, impact statement, abstract and digest of eLife papers (July–Sept 2014)?</p>
<div class="caption-text__body">some more details here</div>
Caption Text Captioned Asset
Many features are conserved between the mammalian nephron and planarian protonephridia.
(A) Image of a young adult C. elegans and schematic depicting the twelve pairs of sensory neurons in the anterior amphid individuals carrying a virus with k deleterious mutations at its endemic equilibrium. class carrying the fewest number of deleterious mutations is defined as mutation class k = 0. Inset: variation in the basic reproductive rate of infected individuals (gray histogram) and variation in the net reproductive rate R of infected individuals (brown histogram) resulting from variation in the number of deleterious mutations carried by circulating viruses.
Listed are, for each triplet of cell types, the probabilities of the four topologies for prior odds ; the number of topologies that reach probability for some value of between 10−6 and 102; the non-null topology that has the highest probability over the range of prior odds (if the null topology is the most likely topology for the entire range of prior odds, the topology is marked ‘null’); and the value of highest probability over the range of prior odds; the correct topology and triplet length in the traditional model; and the correct topology and triplet length in the Adolfsson model.
(A) Doxycycline-inducible gam-gfp fusion construct in the E. coli chromosome. Constitutively produced TetR protein represses the PN25tetO promoter, which produces GamGFP upon doxycycline induction. oriC, origin of replication; ter, replication terminus; arrows, directions of transcription. (B) Phage λ assay for end-blocking activity by Mu Gam and GamGFP. Rolling-circle replication of phage λred gam is inhibited by E. coli RecBCD, which causes small plaques of λred gam on wild-type E. coli (Smith, 1983). Mu Gam protein binds and protects DNA ends from RecBCD exonuclease activity (Akroyd and Symonds, 1986) and so is expected to allow rolling-circle replication of λred gam and therefore allow formation of large plaques. (C) λred gam plaques are small on recB+ (WT) and large on recB-deficient cells (recB-). Plaques produced on WT cells carrying gam and gam-gfp are small when Gam and GamGFP proteins are not produced (Uninduced). (D) λred gam produce large plaques on WT cells if Gam or GamGFP are produced (Induced). (E) UV sensitivity of E. coli recB-null mutant compared with recB+(WT), and uninduced gam and gam-gfp carrying cells. WT ( ), recB− (
), recB− ( ), WT GamGFP, (
), WT GamGFP, ( ); WT Gam, (
); WT Gam, ( ). (F) Induction of Gam or GamGFP with 200 ng/ml doxycycline causes UV sensitivity similar to that of recB-null mutant cells, indicating that Gam or GamGFP block RecBCD action on double-stranded DNA ends. WT, SMR14327; recB, SMR8350; WT GamGFP, SMR14334; WT Gam, SMR14333. Representative experiment performed three times with comparable results.
). (F) Induction of Gam or GamGFP with 200 ng/ml doxycycline causes UV sensitivity similar to that of recB-null mutant cells, indicating that Gam or GamGFP block RecBCD action on double-stranded DNA ends. WT, SMR14327; recB, SMR8350; WT GamGFP, SMR14334; WT Gam, SMR14333. Representative experiment performed three times with comparable results.
HTML
<h6 class="caption-text__heading">Many features are conserved between the mammalian nephron and planarian protonephridia.</h6>
<div class="caption-text__body"><p>(A) Image of a young adult C. elegans and schematic depicting the twelve pairs of sensory neurons in the anterior amphid individuals carrying a virus with k deleterious mutations at its endemic equilibrium. class carrying the fewest number of deleterious mutations is defined as mutation class k = 0. Inset: variation in the basic reproductive rate of infected individuals (gray histogram) and variation in the net reproductive rate R of infected individuals (brown histogram) resulting from variation in the number of deleterious mutations carried by circulating viruses.</p><p>Listed are, for each triplet of cell types, the probabilities of the four topologies for prior odds <math id="inf6"><mrow><mfrac><mrow><mi>p</mi><mrow><mo>(</mo><mrow><msub><mi>β</mi><mi>i</mi></msub><mo>=</mo><mn>1</mn></mrow><mo>)</mo></mrow></mrow><mrow><mi>p</mi><mrow><mo>(</mo><mrow><msub><mi>β</mi><mi>i</mi></msub><mo>=</mo><mn>0</mn></mrow><mo>)</mo></mrow></mrow></mfrac><mo>=</mo><mn>0.05</mn></mrow></math>; the number of topologies that reach probability <math id="inf7"><mstyle displaystyle="true" scriptlevel="0"><mrow><mi>p</mi><mrow><mo>(</mo><mrow><mi>T</mi><mtext> </mtext><mrow></mrow><mo>|</mo><mrow></mrow><mtext> </mtext><mrow><mo>{</mo><msubsup><mi>g</mi><mrow><mi>i</mi></mrow><mrow><mi>A</mi><mo>,</mo><mi>B</mi><mo>,</mo><mi>C</mi></mrow></msubsup><mo>}</mo></mrow></mrow><mo>)</mo></mrow><mo>></mo><mn>0.6</mn></mrow></mstyle></math> for some value of <math id="inf8"><mrow><mfrac><mrow><mi>p</mi><mrow><mo>(</mo><mrow><msub><mi>β</mi><mi>i</mi></msub><mo>=</mo><mn>1</mn></mrow><mo>)</mo></mrow></mrow><mrow><mi>p</mi><mrow><mo>(</mo><mrow><msub><mi>β</mi><mi>i</mi></msub><mo>=</mo><mn>0</mn></mrow><mo>)</mo></mrow></mrow></mfrac><mo> </mo></mrow></math> between 10<sup>−6</sup> and 10<sup>2</sup>; the non-null topology that has the highest probability <math id="inf9"><mstyle displaystyle="true" scriptlevel="0"><mrow><mi>p</mi><mrow><mo>(</mo><mrow><mi>T</mi><mtext> </mtext><mrow></mrow><mo>|</mo><mrow></mrow><mtext> </mtext><mrow><mo>{</mo><msubsup><mi>g</mi><mrow><mi>i</mi></mrow><mrow><mi>A</mi><mo>,</mo><mi>B</mi><mo>,</mo><mi>C</mi></mrow></msubsup><mo>}</mo></mrow></mrow><mo>)</mo></mrow></mrow></mstyle></math> over the range of prior odds (if the null topology is the most likely topology for the entire range of prior odds, the topology is marked ‘null’); and the value of highest probability <math id="inf10"><mstyle displaystyle="true" scriptlevel="0"><mrow><mi>p</mi><mrow><mo>(</mo><mrow><mi>T</mi><mtext> </mtext><mrow></mrow><mo>|</mo><mrow></mrow><mtext> </mtext><mrow><mo>{</mo><msubsup><mi>g</mi><mrow><mi>i</mi></mrow><mrow><mi>A</mi><mo>,</mo><mi>B</mi><mo>,</mo><mi>C</mi></mrow></msubsup><mo>}</mo></mrow></mrow><mo>)</mo></mrow></mrow></mstyle></math> over the range of prior odds; the correct topology and triplet length in the traditional model; and the correct topology and triplet length in the Adolfsson model.</p><p>(<b>A</b>) Doxycycline-inducible <i>gam-gfp</i> fusion construct in the <i>E. coli</i> chromosome. Constitutively produced TetR protein represses the P<sub>N25<i>tetO</i></sub> promoter, which produces GamGFP upon doxycycline induction. <i>oriC</i>, origin of replication; <i>ter</i>, replication terminus; arrows, directions of transcription. (<b>B</b>) Phage λ assay for end-blocking activity by Mu Gam and GamGFP. Rolling-circle replication of phage λ<i>red gam</i> is inhibited by <i>E. coli</i> RecBCD, which causes small plaques of λ<i>red gam</i> on wild-type <i>E. coli</i> (<a href="#">Smith, 1983</a>). Mu Gam protein binds and protects DNA ends from RecBCD exonuclease activity (<a href="#">Akroyd and Symonds, 1986</a>) and so is expected to allow rolling-circle replication of λ<i>red gam</i> and therefore allow formation of large plaques. (<b>C</b>) λ<i>red gam</i> plaques are small on <i>recB</i><sup><i>+</i></sup> (WT) and large on <i>recB</i>-deficient cells (<i>recB</i><sup><i>-</i></sup>). Plaques produced on WT cells carrying <i>gam</i> and <i>gam-gfp</i> are small when Gam and GamGFP proteins are not produced (Uninduced). (<b>D</b>) λ<i>red gam</i> produce large plaques on WT cells if Gam or GamGFP are produced (Induced). (<b>E</b>) UV sensitivity of <i>E</i><i>. coli recB</i>-null mutant compared with <i>recB</i><sup><i>+</i></sup>(WT), and uninduced <i>gam</i> and <i>gam-gfp</i> carrying cells. WT (<img src="https://cdn.elifesciences.org/articles/01222/elife-01222-inf1-v1.jpg">), <i>recB</i><sup>−</sup> (<img src="https://cdn.elifesciences.org/articles/01222/elife-01222-inf2-v1.jpg">), WT GamGFP, (<img src="https://cdn.elifesciences.org/articles/01222/elife-01222-inf3-v1.jpg">); WT Gam, (<img src="https://cdn.elifesciences.org/articles/01222/elife-01222-inf4-v1.jpg">). (<b>F</b>) Induction of Gam or GamGFP with 200 ng/ml doxycycline causes UV sensitivity similar to that of <i>recB-</i>null mutant cells, indicating that Gam or GamGFP block RecBCD action on double-stranded DNA ends. WT, SMR14327; <i>recB</i>, SMR8350; WT GamGFP, SMR14334; WT Gam, SMR14333. Representative experiment performed three times with comparable results.</p></div>
Code
<dock_design>
<SCOREFXNS>
<fullatom weights=beta symmetric = 0> </fullatom>
</SCOREFXNS>
<!-- Longabadaliasametatquedoloreaqueesthicillabadaliasametatquedoloreaqueesthicillabadaliasametatquedoloreaqueesthicillolong -->
<FILTERS>
<Ddg name=ddg scorefxn=fullatom threshold = 0 jump = 1 repeats = 1 repack = 1 confidence = 1/>
<Sasa name=sasa confidence = 0/>
<ShapeComplementarity name=shape verbose = 1 confidence = 0 jump = 1/>
</FILTERS>
<MOVERS>
<AtomTree name=docking_tree docking_ft = 1/>
<DockSetupMover name=setup_dock/>
<DockingProtocol name=dock docking_score_high=fullatom low_res_protocol_only = 0 docking_local_refine = 0 dock_min = 1 />
</MOVERS>
<APPLY_TO_POSE>
</APPLY_TO_POSE>
<PROTOCOLS>
<Add mover_name=docking_tree/>
<Add mover_name=setup_dock/>
<Add mover_name=dock/>
<Add filter_name=ddg/>
<Add filter_name=sasa/>
<Add filter_name=shape/>
</PROTOCOLS>
</dock_design>HTML
<pre class="code"><code>&lt;dock_design&gt;
&nbsp;&nbsp;&lt;SCOREFXNS&gt;
&nbsp;&nbsp;&nbsp;&nbsp;&lt;fullatom weights=beta symmetric = 0&gt; &lt;/fullatom&gt;
&nbsp;&nbsp;&lt;/SCOREFXNS&gt;
&nbsp;&nbsp;&lt;!-- Longabadaliasametatquedoloreaqueesthicillabadaliasametatquedoloreaqueesthicillabadaliasametatquedoloreaqueesthicillolong --&gt;
&nbsp;&nbsp;&lt;FILTERS&gt;
&nbsp;&nbsp;&nbsp;&nbsp;&lt;Ddg name=ddg scorefxn=fullatom threshold = 0 jump = 1 repeats = 1 repack = 1 confidence = 1/&gt;
&nbsp;&nbsp;&nbsp;&nbsp;&lt;Sasa name=sasa confidence = 0/&gt;
&nbsp;&nbsp;&nbsp;&nbsp;&lt;ShapeComplementarity name=shape verbose = 1 confidence = 0 jump = 1/&gt;
&nbsp;&nbsp;&lt;/FILTERS&gt;
&nbsp;&nbsp;&lt;MOVERS&gt;
&nbsp;&nbsp;&nbsp;&nbsp;&lt;AtomTree name=docking_tree docking_ft = 1/&gt;
&nbsp;&nbsp;&nbsp;&nbsp;&lt;DockSetupMover name=setup_dock/&gt;
&nbsp;&nbsp;&nbsp;&nbsp;&lt;DockingProtocol name=dock docking_score_high=fullatom low_res_protocol_only = 0 docking_local_refine = 0 dock_min = 1 /&gt;
&nbsp;&nbsp;&lt;/MOVERS&gt;
&nbsp;&nbsp;&lt;APPLY_TO_POSE&gt;
&nbsp;&nbsp;&lt;/APPLY_TO_POSE&gt;
&nbsp;&nbsp;&lt;PROTOCOLS&gt;
&nbsp;&nbsp;&nbsp;&nbsp;&lt;Add mover_name=docking_tree/&gt;
&nbsp;&nbsp;&nbsp;&nbsp;&lt;Add mover_name=setup_dock/&gt;
&nbsp;&nbsp;&nbsp;&nbsp;&lt;Add mover_name=dock/&gt;
&nbsp;&nbsp;&nbsp;&nbsp;&lt;Add filter_name=ddg/&gt;
&nbsp;&nbsp;&nbsp;&nbsp;&lt;Add filter_name=sasa/&gt;
&nbsp;&nbsp;&nbsp;&nbsp;&lt;Add filter_name=shape/&gt;
&nbsp;&nbsp;&lt;/PROTOCOLS&gt;
&lt;/dock_design&gt;</code></pre>
Date
HTML
<span class="date"> <time datetime="2016-02-29">Feb 29, 2016</time></span>
Date Expanded
HTML
<time class="date date--expanded" datetime="2015-12-18"> <span class="date__prominent">18</span> <span class="date__normal">Dec 2015</span></time>
Date Updated
HTML
<span class="date"> Updated <time datetime="2016-02-29">Feb 29, 2016</time></span>
Definition List
- Research focus
- Pre-mRNA splicing
- post-transcriptional gene regulation
- Experimental organism
- Human
- Mouse
HTML
<dl class="definition-list">
<dt>Research focus</dt><dd>Pre-mRNA splicing</dd><dd>post-transcriptional gene regulation</dd>
<dt>Experimental organism</dt><dd>Human</dd><dd>Mouse</dd>
</dl>
Definition List Inline
- Research focus
- Pre-mRNA splicing
- post-transcriptional gene regulation
- Experimental organism
- Human
- Mouse
HTML
<dl class="definition-list definition-list--inline">
<dt>Research focus</dt><dd>Pre-mRNA splicing</dd><dd>post-transcriptional gene regulation</dd>
<dt>Experimental organism</dt><dd>Human</dd><dd>Mouse</dd>
</dl>
Definition List Timeline With Version
- Version of Record
- Read the peer reviews
- Reviewed Preprint
- v3
- Reviewed Preprint
- v2
- Reviewed Preprint
- v1
HTML
<dl class="definition-list definition-list--timeline" aria-label="Version history">
<dt class="definition-list--vor definition-list--active">Version of Record</dt><dd><time datetime="2023-09-07">September 7, 2023</time><a href="#" aria-label="Descriptive and accessible name">Read the peer reviews</a></dd>
<dt><a href="#" aria-label="Descriptive and accessible name">Reviewed Preprint</a></dt><dd><span class="version">v3</span><time datetime="2023-08-19">August 19, 2023</time></dd>
<dt><a href="#" aria-label="Descriptive and accessible name">Reviewed Preprint</a></dt><dd><span class="version">v2</span><time datetime="2023-05-11">May 11, 2023</time></dd>
<dt><a href="#" aria-label="Descriptive and accessible name">Reviewed Preprint</a></dt><dd><span class="version">v1</span><time datetime="2023-03-19">March 19, 2023</time></dd>
</dl>
Definition List Timeline Without Version
- Version of Record
- Accepted Manuscript
HTML
<dl class="definition-list definition-list--timeline" aria-label="Version history">
<dt class="definition-list--active">Version of Record</dt><dd><time datetime="2023-09-27">September 27, 2023</time></dd>
<dt><a href="#" aria-label="Descriptive and accessible name">Accepted Manuscript</a></dt><dd><time datetime="2023-03-19">March 19, 2023</time></dd>
</dl>
Doi
HTML
<span class="doi">
<a href="https://doi.org/10.7554/eLife.16370" class="doi__link">
https://doi.org/10.7554/eLife.16370
</a>
</span>
Doi Article Section
HTML
<span class="doi doi--article-section">
<a href="https://doi.org/10.7554/eLife.16370" class="doi__link">
https://doi.org/10.7554/eLife.16370
</a>
</span>
Doi Asset
HTML
<span class="doi doi--asset">
<a href="https://doi.org/10.7554/eLife.16370" class="doi__link">
https://doi.org/10.7554/eLife.16370
</a>
</span>
Doi Truncated
HTML
<span class="doi">doi:
<a href="https://doi.org/10.7554/eLife.16370" class="doi__link">
10.7554/eLife.16370
</a>
</span>
Doi Without Link
HTML
<span class="doi">
https://doi.org/10.7554/eLife.16370
</span>
Iframe
HTML
<div class="iframe" style="padding-bottom: 56.25%">
<iframe src="https://www.youtube.com/embed/6j0jRXPvpCE" ></iframe>
</div>
Iframe Figshare
HTML
<div class="iframe" style="padding-bottom: 75%">
<iframe src="https://widgets.figshare.com/articles/8210360/embed" allowfullscreen></iframe>
</div>
Iframe Google Map
HTML
<div class="iframe" style="padding-bottom: 75%">
<iframe src="https://www.google.com/maps/d/embed?mid=13cEQIsP3F9-iEVDDgradCs2Z9F-ODLyx" ></iframe>
</div>
Iframe With Title
HTML
<div class="iframe" style="padding-bottom: 56.25%">
<iframe src="https://www.youtube.com/embed/6j0jRXPvpCE" title="What makes eLife paper webinar" ></iframe>
</div>
Image Link
HTML
<div class="image-link">
<a href="#" class="image-link__link">
<picture class="image-link__picture">
<img src="https://unsplash.it/180/60/"
srcset="https://unsplash.it/360/120/ 2x, https://unsplash.it/180/60/ 1x"
class="image-link__img"
alt="Meaningful alt text here please."
>
</picture>
</a>
</div>
Info Bar
HTML
<div class="info-bar info-bar--info"
>
<div class="info-bar__container">
<div class="info-bar__text">You may want to consider this</div>
</div>
</div>
Info Bar Announcement
HTML
<div class="info-bar info-bar--announcement"
>
<div class="info-bar__container">
<div class="info-bar__text">Very important announcement</div>
</div>
</div>
Info Bar Attention
HTML
<div class="info-bar info-bar--attention"
>
<div class="info-bar__container">
<div class="info-bar__text">Please complete the following tasks</div>
</div>
</div>
Info Bar Correction Notice
HTML
<div class="info-bar info-bar--correction"
>
<div class="info-bar__container">
<div class="info-bar__text">This is a correction notice; read the corrected article <a href="#">Article</a></div>
</div>
</div>
Info Bar Dismissible
HTML
<div class="info-bar info-bar--dismissible" id="12345"
data-behaviour="InfoBar"
data-cookie-name-root="infoBar_"
data-cookie-expires="Tue, 19 January 2038 03:14:07 UTC"
>
<div class="info-bar__container">
<div class="info-bar__text"><a href="#">Read the call for papers</a> for the eLife Special Issue on aging, Geroscience and Longevity.</div>
</div>
</div>
Info Bar Multiple Versions
HTML
<div class="info-bar info-bar--multiple-versions"
>
<div class="info-bar__container">
<div class="info-bar__text">Read the most recent version of this <a href="#">article</a></div>
</div>
</div>
Info Bar Success
HTML
<div class="info-bar info-bar--success"
>
<div class="info-bar__container">
<div class="info-bar__text">Operation completed successfully</div>
</div>
</div>
Info Bar Warning
HTML
<div class="info-bar info-bar--warning"
>
<div class="info-bar__container">
<div class="info-bar__text">Watch out for <a href="#">the thing</a></div>
</div>
</div>
List Heading
Podcasts
HTML
<h3 class="list-heading">Podcasts</h3>
List
- item 1
- item 2
HTML
<ul class="list">
<li>item 1</li>
<li>item 2</li>
</ul>
List Bullet
- item 1
- item 2
HTML
<ul class="list list--bullet">
<li>item 1</li>
<li>item 2</li>
</ul>
List Line
- Version of Record published
- Accepted Manuscript published
- Accepted
- Received
- Preprint posted
HTML
<ul class="list list--line">
<li><strong>Version of Record published</strong><time datetime="2023-09-27">September 27, 2023</time></li>
<li><strong>Accepted Manuscript published</strong><time datetime="2023-09-27">September 27, 2023</time></li>
<li><strong>Accepted</strong><time datetime="2023-09-27">September 27, 2023</time></li>
<li><strong>Received</strong><time datetime="2023-09-27">September 27, 2023</time></li>
<li><strong>Preprint posted</strong><time datetime="2023-09-27">September 27, 2023</time></li>
</ul>
List Numbers
- item 1
- item 2
HTML
<ol class="list list--number">
<li>item 1</li>
<li>item 2</li>
</ol>
List Roman
- item 1
- item 2
HTML
<ol class="list list--roman-lower">
<li>item 1</li>
<li>item 2</li>
</ol>
Math
HTML
<div class="math-block" >
<span class="math-block__math"><math><mrow><mtext>Body Mass</mtext><mo>=</mo><mn>0.060</mn><mo>×</mo><mtext>FSTpr</mtext><mo>+</mo><mn>13</mn><mo>.</mo><mn>856</mn><mo>,</mo><mtext>SEE</mtext><mo>=</mo><mn>6</mn><mo>.</mo><mn>78</mn><mo>,</mo><mtext>r</mtext><mo>=</mo><mn>0</mn><mo>.</mo><mn>50</mn><mo>,</mo><mtext>p</mtext><mo>=</mo><mo><</mo><mn>0</mn><mo>.</mo><mn>001</mn><mo>.</mo></mrow></math></span>
</div>
Math Long
HTML
<div class="math-block" id="equ7">
<span class="math-block__label">(7)</span>
<span class="math-block__math"><math><mstyle displaystyle="true" scriptlevel="0"><mrow><mtable columnalign="left left" columnspacing="1em" rowspacing="4pt"><mtr><mtd><mi>y</mi><mo>=</mo><msub><mi>β</mi><mrow><mn>0</mn></mrow></msub><mo>+</mo><msub><mi>β</mi><mrow><mn>1</mn></mrow></msub><mspace width="thinmathspace"/><mi>C</mi><mi>h</mi><mi>o</mi><mi>i</mi><mi>c</mi><mi>e</mi><mo>+</mo><msub><mi>β</mi><mrow><mn>2</mn></mrow></msub><mspace width="thinmathspace"/><mi>S</mi><mi>V</mi><mi>s</mi><mi>p</mi><mi>e</mi><mi>n</mi><mi>d</mi><mo>+</mo><msub><mi>β</mi><mrow><mn>3</mn></mrow></msub><mspace width="thinmathspace"/><mi>S</mi><mi>V</mi><mi>s</mi><mi>a</mi><mi>v</mi><mi>e</mi><mo>+</mo><msub><mi>β</mi><mrow><mn>4</mn></mrow></msub><mspace width="thinmathspace"/><mi>S</mi><mi>e</mi><mi>q</mi><mi>S</mi><mi>V</mi><mo>+</mo><msub><mi>β</mi><mrow><mn>5</mn></mrow></msub><mspace width="thinmathspace"/><mi>S</mi><mi>e</mi><mi>q</mi><mi>L</mi><mi>e</mi><mi>n</mi><mi>g</mi><mi>t</mi><mi>h</mi><mo>+</mo><msub><mi>β</mi><mrow><mn>6</mn></mrow></msub><mspace width="thinmathspace"/><mi>C</mi><mi>u</mi><mi>e</mi><mspace width="thinmathspace"/><mi>p</mi><mi>o</mi><mi>s</mi><mi>i</mi><mi>t</mi><mi>i</mi><mi>o</mi><mi>n</mi><mo>+</mo></mtd></mtr><mtr><mtd><msub><mi>β</mi><mrow><mn>7</mn></mrow></msub><mspace width="thinmathspace"/><mi>L</mi><mi>e</mi><mi>f</mi><mi>t</mi><mrow><mo>/</mo></mrow><mi>r</mi><mi>i</mi><mi>g</mi><mi>h</mi><mi>t</mi><mo>+</mo><msub><mi>β</mi><mrow><mn>8</mn></mrow></msub><mspace width="thinmathspace"/><mi>S</mi><mi>e</mi><mi>q</mi><mi>u</mi><mi>e</mi><mi>n</mi><mi>c</mi><mi>e</mi><mspace width="thinmathspace"/><mi>p</mi><mi>r</mi><mi>o</mi><mi>g</mi><mi>r</mi><mi>e</mi><mi>s</mi><mi>s</mi><mo>+</mo><mi>ϵ</mi><mo>,</mo></mtd></mtr></mtable></mrow></mstyle></math></span>
</div>
Math With Label
HTML
<div class="math-block" id="equ10">
<span class="math-block__label">(10)</span>
<span class="math-block__math"><math><mstyle displaystyle="true" scriptlevel="0"><mrow><mi>S</mi><mi>V</mi><mi>s</mi><mi>a</mi><mi>v</mi><msub><mi>e</mi><mrow><mi>n</mi></mrow></msub><mo>=</mo><mtext> </mtext><mfrac><mn>1</mn><mrow><mi>m</mi><mo>−</mo><mi>n</mi></mrow></mfrac><munderover><mo movablelimits="false">∑</mo><mrow><mi>i</mi><mo>=</mo><mi>n</mi><mo>+</mo><mn>1</mn></mrow><mrow><mi>m</mi></mrow></munderover><mi>S</mi><mi>V</mi><mi>s</mi><mi>p</mi><mi>e</mi><mi>n</mi><msub><mi>d</mi><mrow><mi>i</mi></mrow></msub><mo>,</mo></mrow></mstyle></math></span>
</div>
Media Source
HTML
<source src="#uri-of-a-media-source" type='video/mp4' class="media-source"/>
<div>
<a href="#uri-of-a-media-source" class="media-source__fallback_link"
>download <b>amazing</b> artefact (mp3)</a>
</div>
Message Bar
HTML
<div class="message-bar">
1824 results found
</div>
Modal Window
HTML
<div class="modal modal-nojs" data-behaviour="Modal">
<a href="" class="modal-click">Open modal window</a>
<div class="modal-container">
<div class="modal-content">
<h6>This is a title</h6>
<a href="" class="modal-content__button modal-content__close-button">Close</a>
<div class="modal-content__body">
This is some body text
</div>
</div>
</div>
</div>
Modal Window Citations
Cite this article
Close- Schrenk F
- Bromage TG
- Betzler CG
- Ring U
- Juwayeyi YM
- Schrenk F
- Bromage TG
- Betzler CG
- Ring U
- Juwayeyi YM
- Schrenk F
- Bromage TG
- Betzler CG
- Ring U
- Juwayeyi YM
- Schrenk F
- Bromage TG
- Betzler CG
- Ring U
- Juwayeyi YM
- Schrenk F
- Bromage TG
- Betzler CG
- Ring U
- Juwayeyi YM
HTML
<div class="modal modal-nojs" data-behaviour="Modal">
<a href="" class="modal-click">Open modal window</a>
<div class="modal-container">
<div class="modal-content">
<h6>Cite this article</h6>
<a href="" class="modal-content__button modal-content__close-button">Close</a>
<div class="modal-content__body">
<div class='reference'><ol class='reference__authors_list'><li class='reference__author'><a href='https://scholar.google.com/scholar?q=%22author:Schrenk%20F%22' class='reference__authors_link'>Schrenk F</a></li><li class='reference__author'> Bromage TG</li><li class='reference__author'> Betzler CG</li><li class='reference__author'> Ring U</li><li class='reference__author'> Juwayeyi YM</li><li class='reference__author'> <a href='https://scholar.google.com/scholar?q=%22author:Schrenk%20F%22' class='reference__authors_link'>Schrenk F</a></li><li class='reference__author'> Bromage TG</li><li class='reference__author'> Betzler CG</li><li class='reference__author'> Ring U</li><li class='reference__author'> Juwayeyi YM</li><li class='reference__author'> <a href='https://scholar.google.com/scholar?q=%22author:Schrenk%20F%22' class='reference__authors_link'>Schrenk F</a></li><li class='reference__author'> Bromage TG</li><li class='reference__author'> Betzler CG</li><li class='reference__author'> Ring U</li><li class='reference__author'> Juwayeyi YM</li><li class='reference__author'> <a href='https://scholar.google.com/scholar?q=%22author:Schrenk%20F%22' class='reference__authors_link'>Schrenk F</a></li><li class='reference__author'> Bromage TG</li><li class='reference__author'> Betzler CG</li><li class='reference__author'> Ring U</li><li class='reference__author'> Juwayeyi YM</li><li class='reference__author'> <a href='https://scholar.google.com/scholar?q=%22author:Schrenk%20F%22' class='reference__authors_link'>Schrenk F</a></li><li class='reference__author'> Bromage TG</li><li class='reference__author'> Betzler CG</li><li class='reference__author'> Ring U</li><li class='reference__author'>Juwayeyi YM</li></ol> <span class='reference__authors_list_suffix'>(1993)</span> <div class='reference__title'>Oldest <i>Homo</i> and Pliocene biogeography of the Malawi rift</div><div class='reference__origin'> <i>Nature</i> <strong>365</strong>:833–836.</div><span class='doi'>https://doi.org/10.1038/365833a0.1</span></div><ol class='button-collection'><li class='button-collection__item'> <button class='button button--default' type='button' data-behaviour='ButtonClipboard' data-clipboard='Schrenk F Bromage TG Betzler CG Ring U Juwayeyi YM Schrenk F Bromage TG Betzler CG Ring U Juwayeyi YM Schrenk F Bromage TG Betzler CG Ring U Juwayeyi YM Schrenk F Bromage TG Betzler CG Ring U Juwayeyi YM Schrenk F Bromage TG Betzler CG Ring UJuwayeyi YM(1993) Oldest Homo and Pliocene biogeography of the Malawi rift Nature 365:833–836.https://doi.org/10.1038/365833a0.1'>Copy to clipboard</button></li><li class='button-collection__item'> <button class='button button--default button--secondary' type='button'>Download Bibtex</button></li><li class='button-collection__item'> <button class='button button--default button--secondary' type='button'>Download .RIS</button></li></ol>
</div>
</div>
</div>
</div>
Modal Window Share
HTML
<div class="modal modal-nojs" data-behaviour="Modal">
<a href="" class="modal-click">Open modal window</a>
<div class="modal-container">
<div class="modal-content">
<h6>Share this article</h6>
<a href="" class="modal-content__button modal-content__close-button">Close</a>
<div class="modal-content__body">
<div class='form-item'><input type='url' class='text-field text-field--url' value='https://doi.org/10.7554/eLife.09560.1'></div><button class='button button--default' type='submit' id='button-id' data-behaviour='ButtonClipboard' data-clipboard='https://doi.org/10.7554/eLife.09560.1'>Copy to clipboard</button><ul class='social-media-sharers'><li><a class='social-media-sharer email' href='mailto:?subject=Assessing%20the%20causal%20role%20of%20epigenetic%20clocks%20in%20the%20development%20of%20multiple%20cancers%3A%20a%20Mendelian%20randomization%20study&body=https%3A%2F%2Fdoi.org%2F10.7554%2FeLife.75374' target='_blank' rel='noopener noreferrer' aria-label='Share by Email'><svg width='24px' height='24px' viewBox='0 0 24 24' version='1.1' xmlns='http://www.w3.org/2000/svg' xmlns:xlink='http://www.w3.org/1999/xlink'><g id='email' stroke='none' stroke-width='1' fill='none' fill-rule='evenodd'><rect id='clear-button-bg' x='0' y='0' width='24' height='24'></rect><path d='M20,4 L4,4 C2.9,4 2.01,4.9 2.01,6 L2,18 C2,19.1 2.9,20 4,20 L20,20 C21.1,20 22,19.1 22,18 L22,6 C22,4.9 21.1,4 20,4 Z M20,8 L12,13 L4,8 L4,6 L12,11 L20,6 L20,8 Z' id='Shape' fill='#000000' fill-rule='nonzero'></path></g></svg></a></li><li><a class='social-media-sharer' href='https://twitter.com/intent/tweet/?text=In%20%40eLife%3A%20Assessing%20the%20causal%20role%20of%20epigenetic%20clocks%20in%20the%20development%20of%20multiple%20cancers%3A%20a%20Mendelian%20randomization%20study&url=https%3A%2F%2Fdoi.org%2F10.7554%2FeLife.75374' target='_blank' rel='noopener noreferrer' aria-label='Tweet a link to this page'><svg width='24px' height='24px' viewBox='0 0 24 24' version='1.1' xmlns='http://www.w3.org/2000/svg' xmlns:xlink='http://www.w3.org/1999/xlink'><g id='x-24-articles' stroke='none' stroke-width='1' fill='none' fill-rule='evenodd'><rect id='Rectangle' x='0' y='0' width='24' height='24'></rect><path d='M13.6604555,11.4785798 L20.954365,3 L19.225942,3 L12.8926412,10.3618317 L7.83425247,3 L2,3 L9.64927632,14.1323934 L2,23.023486 L3.72852106,23.023486 L10.4166498,15.2491415 L15.7586789,23.023486 L21.5929313,23.023486 L13.660031,11.4785798 L13.6604555,11.4785798 Z M11.293009,14.2304723 L10.5179779,13.1219369 L4.35133136,4.30120576 L7.00623887,4.30120576 L11.9827944,11.4198173 L12.7578255,12.5283527 L19.2267583,21.7814574 L16.5718508,21.7814574 L11.293009,14.2308968 L11.293009,14.2304723 Z' id='Shape' fill='#212121' fill-rule='nonzero'></path></g></svg></a></li><li><a class='social-media-sharer' href='https://facebook.com/sharer/sharer.php?u=https%3A%2F%2Fdoi.org%2F10.7554%2FeLife.75374' target='_blank' rel='noopener noreferrer' aria-label='Share on Facebook'><svg width='24px' height='24px' viewBox='0 0 24 24' version='1.1' xmlns='http://www.w3.org/2000/svg' xmlns:xlink='http://www.w3.org/1999/xlink'><g id='facebook' stroke='none' stroke-width='1' fill='none' fill-rule='evenodd'><g><rect id='clear-button-bg' x='0' y='0' width='24' height='24'></rect><path d='M11.8969072,2 C6.43099227,2 2,6.41067993 2,11.8515383 C2,16.7687258 5.61915593,20.844338 10.3505155,21.5833958 L10.3505155,14.6992486 L7.83762887,14.6992486 L7.83762887,11.8515383 L10.3505155,11.8515383 L10.3505155,9.68112126 C10.3505155,7.21207948 11.8280541,5.84825714 14.0887242,5.84825714 C15.1715271,5.84825714 16.3041237,6.04067 16.3041237,6.04067 L16.3041237,8.465072 L15.0561469,8.465072 C13.8267075,8.465072 13.443299,9.22446783 13.443299,10.0035475 L13.443299,11.8515383 L16.1881443,11.8515383 L15.7493557,14.6992486 L13.443299,14.6992486 L13.443299,21.5833958 C18.1746585,20.844338 21.7938144,16.7687258 21.7938144,11.8515383 C21.7938144,6.41067993 17.3628222,2 11.8969072,2 Z' id='Fill-1' fill='#212121' fill-rule='nonzero'></path></g></g></svg></a></li><li><a class='social-media-sharer' href='https://www.linkedin.com/shareArticle?title=Assessing%20the%20causal%20role%20of%20epigenetic%20clocks%20in%20the%20development%20of%20multiple%20cancers%3A%20a%20Mendelian%20randomization%20study&url=https%3A%2F%2Fdoi.org%2F10.7554%2FeLife.75374' target='_self' aria-label='Share this page to LinkedIn (opens up email program, if configured on this system)'><svg width='24px' height='24px' viewBox='0 0 24 24' version='1.1' xmlns='http://www.w3.org/2000/svg' xmlns:xlink='http://www.w3.org/1999/xlink'><g id='linkedin' stroke='none' stroke-width='1' fill='none' fill-rule='evenodd'><g><rect id='Rectangle' x='0' y='0' width='24' height='24'></rect><path d='M18.9751954,18.9751435 L16.0232549,18.9751435 L16.0232549,14.3522816 C16.0232549,13.2499163 16.0035753,11.8308324 14.4879383,11.8308324 C12.9504693,11.8308324 12.7152366,13.0319032 12.7152366,14.2720257 L12.7152366,18.974836 L9.76329605,18.974836 L9.76329605,9.46835755 L12.5971589,9.46835755 L12.5971589,10.7675189 L12.6368256,10.7675189 C13.2146663,9.77952684 14.2890899,9.18943873 15.4328668,9.23189481 C18.4247815,9.23189481 18.9764254,11.1998552 18.9764254,13.7600486 L18.9751954,18.9751435 Z M6.43252317,8.16888879 C5.48643292,8.16905854 4.71933758,7.40223852 4.7191677,6.45614827 C4.71899794,5.51005802 5.48581795,4.74296266 6.4319082,4.74279278 C7.37799845,4.74262301 8.14509382,5.50944301 8.14526371,6.45553326 C8.14534522,6.90986184 7.96494211,7.34561469 7.64374096,7.66693118 C7.32253981,7.98824766 6.88685175,8.16880719 6.43252317,8.16888879 M7.90849343,18.9751435 L4.95347798,18.9751435 L4.95347798,9.46835755 L7.90849343,9.46835755 L7.90849343,18.9751435 Z M20.4468607,2.00148552 L3.47012787,2.00148552 C2.66777241,1.99243094 2.00979231,2.63513479 2,3.43748159 L2,20.4846305 C2.00945705,21.2873639 2.6673842,21.9307041 3.47012787,21.9223954 L20.4468607,21.9223954 C21.2511925,21.9322515 21.9116962,21.2889485 21.922831,20.4846305 L21.922831,3.43625161 C21.9113617,2.63231978 21.2508045,1.98965267 20.4468607,2' id='Path_2520' fill='#212121' fill-rule='nonzero'></path></g></g></svg></a></li><li><a class='social-media-sharer' href='https://reddit.com/submit/?title=Assessing%20the%20causal%20role%20of%20epigenetic%20clocks%20in%20the%20development%20of%20multiple%20cancers%3A%20a%20Mendelian%20randomization%20study&url=https%3A%2F%2Fdoi.org%2F10.7554%2FeLife.75374' target='_blank' rel='noopener noreferrer' aria-label='Share this page on Reddit'><svg width='24px' height='24px' viewBox='0 0 24 24' version='1.1' xmlns='http://www.w3.org/2000/svg' xmlns:xlink='http://www.w3.org/1999/xlink'><g id='reddit' stroke='none' stroke-width='1' fill='none' fill-rule='evenodd'><g><rect id='Rectangle' x='0' y='0' width='24' height='24'></rect><path d='M11.9872095,1.051163 C18.0270212,1.051163 22.923256,5.94739779 22.923256,11.9872095 C22.923256,18.0270212 18.0270212,22.923256 11.9872095,22.923256 C5.94739779,22.923256 1.051163,18.0270212 1.051163,11.9872095 C1.051163,5.94739779 5.94739779,1.051163 11.9872095,1.051163 Z M16.5534886,5.37441881 C16.1058142,5.37441881 15.7220932,5.63023277 15.5430235,6.0139537 L12.895349,5.451163 C12.8186049,5.4383723 12.7418607,5.451163 12.6779072,5.48953509 C12.6139537,5.52790719 12.5755816,5.59186067 12.5500002,5.66860486 L11.7441863,9.48023277 C10.0430235,9.53139556 8.52093044,10.0302328 7.42093044,10.8360467 C7.13953509,10.5674421 6.74302347,10.3883723 6.32093044,10.3883723 C5.4383723,10.3883723 4.72209323,11.1046514 4.72209323,11.9872095 C4.72209323,12.6395351 5.10581416,13.1895351 5.66860486,13.445349 C5.64302347,13.5988374 5.63023277,13.7651165 5.63023277,13.9313956 C5.63023277,16.3872095 8.48255835,18.3697677 12.0127909,18.3697677 C15.5430235,18.3697677 18.395349,16.3872095 18.395349,13.9313956 C18.395349,13.7651165 18.3825583,13.6116281 18.356977,13.4581397 C18.8813956,13.2023258 19.2779072,12.6395351 19.2779072,11.9872095 C19.2779072,11.1046514 18.5616281,10.3883723 17.67907,10.3883723 C17.2441863,10.3883723 16.8604653,10.5546514 16.57907,10.8360467 C15.4918607,10.0558142 13.9825583,9.54418626 12.3197677,9.48023277 L13.0488374,6.06511649 L15.4151165,6.5639537 C15.4406979,7.16511649 15.9395351,7.651163 16.5534886,7.651163 C17.1802328,7.651163 17.6918607,7.13953509 17.6918607,6.51279091 C17.6918607,5.88604672 17.1802328,5.37441881 16.5534886,5.37441881 Z M9.72325602,15.7093025 C10.2093025,16.195349 11.2581397,16.3744188 12.0127909,16.3744188 C12.7674421,16.3744188 13.8034886,16.195349 14.3023258,15.7093025 C14.4174421,15.5941863 14.6093025,15.5941863 14.7244188,15.7093025 C14.8139537,15.8372095 14.8139537,16.0162793 14.6988374,16.1313956 C13.9186049,16.9116281 12.4348839,16.9627909 12.0000002,16.9627909 C11.5651165,16.9627909 10.0686049,16.8988374 9.301163,16.1313956 C9.18604672,16.0162793 9.18604672,15.8244188 9.301163,15.7093025 C9.41627928,15.5941863 9.60813974,15.5941863 9.72325602,15.7093025 Z M9.48023277,11.9872095 C10.1069769,11.9872095 10.6186049,12.4988374 10.6186049,13.1255816 C10.6186049,13.7523258 10.1069769,14.2639537 9.48023277,14.2639537 C8.85348858,14.2639537 8.34186067,13.7523258 8.34186067,13.1255816 C8.34186067,12.4988374 8.85348858,11.9872095 9.48023277,11.9872095 Z M14.4941863,11.9872095 C15.1209304,11.9872095 15.6325583,12.4988374 15.6325583,13.1255816 C15.6325583,13.7523258 15.1209304,14.2639537 14.4941863,14.2639537 C13.8674421,14.2639537 13.3558142,13.7523258 13.3558142,13.1255816 C13.3558142,12.4988374 13.8674421,11.9872095 14.4941863,11.9872095 Z' id='Combined-Shape' fill='#212121' fill-rule='nonzero'></path></g></g></svg></a></li><li><a class='social-media-sharer' href='https://toot.kytta.dev/?text=Assessing%20the%20causal%20role%20of%20epigenetic%20clocks%20in%20the%20development%20of%20multiple%20cancers%3A%20a%20Mendelian%20randomization%20study%20https%3A%2F%2Fdoi.org%2F10.7554%2FeLife.75374' target='_blank' rel='noopener noreferrer' aria-label='Share this page on Mastodon'><svg width='24px' height='24px' viewBox='0 0 24 24' version='1.1' xmlns='http://www.w3.org/2000/svg' xmlns:xlink='http://www.w3.org/1999/xlink'><g id='mastodon' stroke='none' stroke-width='1' fill='none' fill-rule='evenodd'><g><rect id='Rectangle' x='0' y='0' width='24' height='24'></rect><path d='M11.5412949,0 C14.6936837,0.0250851513 17.7279692,0.357269448 19.4950021,1.1471931 C19.4950021,1.1471931 22.9995752,2.67324958 22.9995752,7.87958222 L23,8.27840764 C22.9948717,9.32900538 22.9399626,12.2394095 22.5108214,14.3877566 C22.1727192,16.0807457 19.4826471,17.9335557 16.3930961,18.2926354 C14.7820287,18.4797396 13.1958041,18.6517151 11.5043627,18.5762011 C8.73816776,18.4528442 6.55544838,17.9335557 6.55544838,17.9335557 C6.55544838,18.1956567 6.57205459,18.4452152 6.605267,18.6786105 C6.96489093,21.335697 9.312211,21.4948713 11.5357152,21.5690923 C13.7799439,21.6438306 15.7782681,21.0305374 15.7782681,21.0305374 L15.8704657,23.0052819 C15.8704657,23.0052819 14.3007146,23.8257215 11.5043627,23.9766203 C9.96237703,24.0591169 8.04774824,23.9388633 5.8177344,23.3643616 C0.981210876,22.1183796 0.149439349,17.1004442 0.022169409,12.0089343 C-0.0166226796,10.4972307 0.00729025582,9.07177343 0.00729025582,7.87958222 C0.00729025582,2.67324958 3.51199627,1.1471931 3.51199627,1.1471931 C5.27916201,0.357269448 8.31145476,0.0250851513 11.4638436,0 L11.5412949,0 Z M15.8599153,5 C14.461897,5 13.4033065,5.46697151 12.7035195,6.40103745 L12.0229667,7.39238391 L11.3425554,6.40103745 C10.642627,5.46697151 9.58403654,5 8.18615964,5 C6.97793807,5 6.00462882,5.36912752 5.26142334,6.08919046 C4.54070495,6.8092534 4.18190148,7.78253068 4.18190148,9.00730143 L4.18190148,15 L6.91358821,15 L6.91358821,9.1834452 C6.91358821,7.95732233 7.50716259,7.33498047 8.69445278,7.33498047 C10.0071899,7.33498047 10.6652556,8.0732355 10.6652556,9.5330285 L10.6652556,12.7167687 L13.3808194,12.7167687 L13.3808194,9.5330285 C13.3808194,8.0732355 14.0387436,7.33498047 15.3514807,7.33498047 C16.5387709,7.33498047 17.1323453,7.95732233 17.1323453,9.1834452 L17.1323453,15 L19.864032,15 L19.864032,9.00730143 C19.864032,7.78253068 19.5052285,6.8092534 18.7846516,6.08919046 C18.0413047,5.36912752 17.0679954,5 15.8599153,5 Z' id='Combined-Shape' fill='#212121' fill-rule='nonzero'></path></g></g></svg></a></li></ul>
</div>
</div>
</div>
</div>
Nav Linked Item
atoms-button
HTML
<li class="nav-primary__item nav-primary__item--first">
<a href="#mainMenu">
Menu
</a>
</li>
Orcid

HTML
<span class="orcid">
<a href="https://orcid.org/0000-0002-3400-7927">
<picture>
<source srcset="../../assets/img/icons/orcid.svg" type="image/svg+xml">
<img src="../../assets/img/icons/orcid.png" class="orcid__icon"
alt="ORCID icon">
</picture> <span class="visuallyhidden">"This ORCID iD identifies the author of this article:"</span>
0000-0002-3400-7927</a>
</span>
Paragraph
Some text in a paragraph element
HTML
<p class="paragraph">Some text in a paragraph element</p>
Picture

HTML
<picture>
<source srcset="../../assets/img/patterns/molecules/nav-primary-menu-ic.svg"
type="image/svg+xml"
>
<img srcset="../../assets/img/patterns/molecules/nav-primary-menu-ic_2x.png 2x, ../../assets/img/patterns/molecules/nav-primary-menu-ic_1x.png 1x"
src="../../assets/img/patterns/molecules/nav-primary-menu-ic_1x.png"
alt="Meaningful alt text here please." />
</picture>
Picture Svg With Fallback

HTML
<picture>
<source srcset="../../assets/img/patterns/molecules/nav-primary-menu-ic.svg"
type="image/svg+xml"
>
<img srcset="../../assets/img/patterns/molecules/nav-primary-menu-ic_2x.png 2x, ../../assets/img/patterns/molecules/nav-primary-menu-ic_1x.png 1x"
src="../../assets/img/patterns/molecules/nav-primary-menu-ic_1x.png"
alt="Meaningful alt text here please." />
</picture>
Previous Version Warning
HTML
<div class="previous-version-warning-container">
<div class="previous-version-warning">
<span class="previous-version-warning__text">A newer version is available.</span>
<span class="previous-version-warning__text">
<a class="previous-version-warning__link" href="#">Read the latest version</a>.
</span>
</div>
</div>
Process Block New Process
Version of Record: This is the final version of the article.
Read more about eLife's peer review process.HTML
<div class="process-block">
<div class="process-block__bar process-block__bar-vor"></div>
<div class="process-block__content">
<p><strong>Version of Record: </strong>This is the final version of the article.</p>
<a href="#" class="process-block__link">Read more about eLife's peer review process.</a>
</div>
</div>
Section Listing Link
HTML
<a href="#subjectListing" class="section-listing-link" data-behaviour="SectionListingLink">All sections</a>
See More Link
HTML
<a href="#" class="see-more-link">wonderous things!</a>
See More Link Inline
HTML
<a href="#" class="see-more-link see-more-link--inline" aria-label="wonderous things inline!">wonderous things inline!</a>
Social Media Sharers Journal
HTML
<ul class="social-media-sharers">
<li>
<a class="social-media-sharer email" href="mailto:?subject=Some%20article%20title&amp;body=https%3A%2F%2Fexample.com%2Fsome-article-url" target="_blank" rel="noopener noreferrer" aria-label="Share by Email">
<svg width="24px" height="24px" viewBox="0 0 24 24" version="1.1" xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink">
<g id="email" stroke="none" stroke-width="1" fill="none" fill-rule="evenodd">
<rect id="clear-button-bg" x="0" y="0" width="24" height="24"></rect>
<path d="M20,4 L4,4 C2.9,4 2.01,4.9 2.01,6 L2,18 C2,19.1 2.9,20 4,20 L20,20 C21.1,20 22,19.1 22,18 L22,6 C22,4.9 21.1,4 20,4 Z M20,8 L12,13 L4,8 L4,6 L12,11 L20,6 L20,8 Z" id="Shape" fill="#000000" fill-rule="nonzero"></path>
</g>
</svg>
</a>
</li>
<li>
<a class="social-media-sharer" href="https://twitter.com/intent/tweet/?text=Some%20article%20title&amp;url=https%3A%2F%2Fexample.com%2Fsome-article-url" target="_blank" rel="noopener noreferrer" aria-label="Tweet a link to this page">
<svg width="24px" height="24px" viewBox="0 0 24 24" version="1.1" xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink">
<g id="x-24-articles" stroke="none" stroke-width="1" fill="none" fill-rule="evenodd">
<rect id="Rectangle" x="0" y="0" width="24" height="24"></rect>
<path d="M13.6604555,11.4785798 L20.954365,3 L19.225942,3 L12.8926412,10.3618317 L7.83425247,3 L2,3 L9.64927632,14.1323934 L2,23.023486 L3.72852106,23.023486 L10.4166498,15.2491415 L15.7586789,23.023486 L21.5929313,23.023486 L13.660031,11.4785798 L13.6604555,11.4785798 Z M11.293009,14.2304723 L10.5179779,13.1219369 L4.35133136,4.30120576 L7.00623887,4.30120576 L11.9827944,11.4198173 L12.7578255,12.5283527 L19.2267583,21.7814574 L16.5718508,21.7814574 L11.293009,14.2308968 L11.293009,14.2304723 Z" id="Shape" fill="#212121" fill-rule="nonzero"></path>
</g>
</svg>
</a>
</li>
<li>
<a class="social-media-sharer" href="https://facebook.com/sharer/sharer.php?u=https%3A%2F%2Fexample.com%2Fsome-article-url" target="_blank" rel="noopener noreferrer" aria-label="Share on Facebook">
<svg width="24px" height="24px" viewBox="0 0 24 24" version="1.1" xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink">
<g id="facebook" stroke="none" stroke-width="1" fill="none" fill-rule="evenodd">
<g>
<rect id="clear-button-bg" x="0" y="0" width="24" height="24"></rect>
<path d="M11.8969072,2 C6.43099227,2 2,6.41067993 2,11.8515383 C2,16.7687258 5.61915593,20.844338 10.3505155,21.5833958 L10.3505155,14.6992486 L7.83762887,14.6992486 L7.83762887,11.8515383 L10.3505155,11.8515383 L10.3505155,9.68112126 C10.3505155,7.21207948 11.8280541,5.84825714 14.0887242,5.84825714 C15.1715271,5.84825714 16.3041237,6.04067 16.3041237,6.04067 L16.3041237,8.465072 L15.0561469,8.465072 C13.8267075,8.465072 13.443299,9.22446783 13.443299,10.0035475 L13.443299,11.8515383 L16.1881443,11.8515383 L15.7493557,14.6992486 L13.443299,14.6992486 L13.443299,21.5833958 C18.1746585,20.844338 21.7938144,16.7687258 21.7938144,11.8515383 C21.7938144,6.41067993 17.3628222,2 11.8969072,2 Z" id="Fill-1" fill="#212121" fill-rule="nonzero"></path>
</g>
</g>
</svg>
</a>
</li>
<li>
<a class="social-media-sharer" href="https://www.linkedin.com/shareArticle?url=https%3A%2F%2Fexample.com%2Fsome-article-url" target="_self" aria-label="Share this page to LinkedIn (opens up email program, if configured on this system)">
<svg width="24px" height="24px" viewBox="0 0 24 24" version="1.1" xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink">
<g id="linkedin" stroke="none" stroke-width="1" fill="none" fill-rule="evenodd">
<g>
<rect id="Rectangle" x="0" y="0" width="24" height="24"></rect>
<path d="M18.9751954,18.9751435 L16.0232549,18.9751435 L16.0232549,14.3522816 C16.0232549,13.2499163 16.0035753,11.8308324 14.4879383,11.8308324 C12.9504693,11.8308324 12.7152366,13.0319032 12.7152366,14.2720257 L12.7152366,18.974836 L9.76329605,18.974836 L9.76329605,9.46835755 L12.5971589,9.46835755 L12.5971589,10.7675189 L12.6368256,10.7675189 C13.2146663,9.77952684 14.2890899,9.18943873 15.4328668,9.23189481 C18.4247815,9.23189481 18.9764254,11.1998552 18.9764254,13.7600486 L18.9751954,18.9751435 Z M6.43252317,8.16888879 C5.48643292,8.16905854 4.71933758,7.40223852 4.7191677,6.45614827 C4.71899794,5.51005802 5.48581795,4.74296266 6.4319082,4.74279278 C7.37799845,4.74262301 8.14509382,5.50944301 8.14526371,6.45553326 C8.14534522,6.90986184 7.96494211,7.34561469 7.64374096,7.66693118 C7.32253981,7.98824766 6.88685175,8.16880719 6.43252317,8.16888879 M7.90849343,18.9751435 L4.95347798,18.9751435 L4.95347798,9.46835755 L7.90849343,9.46835755 L7.90849343,18.9751435 Z M20.4468607,2.00148552 L3.47012787,2.00148552 C2.66777241,1.99243094 2.00979231,2.63513479 2,3.43748159 L2,20.4846305 C2.00945705,21.2873639 2.6673842,21.9307041 3.47012787,21.9223954 L20.4468607,21.9223954 C21.2511925,21.9322515 21.9116962,21.2889485 21.922831,20.4846305 L21.922831,3.43625161 C21.9113617,2.63231978 21.2508045,1.98965267 20.4468607,2" id="Path_2520" fill="#212121" fill-rule="nonzero"></path>
</g>
</g>
</svg>
</a>
</li>
<li>
<a class="social-media-sharer" href="https://reddit.com/submit/?url=https%3A%2F%2Fexample.com%2Fsome-article-url" target="_blank" rel="noopener noreferrer" aria-label="Share this page on Reddit">
<svg width="24px" height="24px" viewBox="0 0 24 24" version="1.1" xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink">
<g id="reddit" stroke="none" stroke-width="1" fill="none" fill-rule="evenodd">
<g>
<rect id="Rectangle" x="0" y="0" width="24" height="24"></rect>
<path d="M11.9872095,1.051163 C18.0270212,1.051163 22.923256,5.94739779 22.923256,11.9872095 C22.923256,18.0270212 18.0270212,22.923256 11.9872095,22.923256 C5.94739779,22.923256 1.051163,18.0270212 1.051163,11.9872095 C1.051163,5.94739779 5.94739779,1.051163 11.9872095,1.051163 Z M16.5534886,5.37441881 C16.1058142,5.37441881 15.7220932,5.63023277 15.5430235,6.0139537 L12.895349,5.451163 C12.8186049,5.4383723 12.7418607,5.451163 12.6779072,5.48953509 C12.6139537,5.52790719 12.5755816,5.59186067 12.5500002,5.66860486 L11.7441863,9.48023277 C10.0430235,9.53139556 8.52093044,10.0302328 7.42093044,10.8360467 C7.13953509,10.5674421 6.74302347,10.3883723 6.32093044,10.3883723 C5.4383723,10.3883723 4.72209323,11.1046514 4.72209323,11.9872095 C4.72209323,12.6395351 5.10581416,13.1895351 5.66860486,13.445349 C5.64302347,13.5988374 5.63023277,13.7651165 5.63023277,13.9313956 C5.63023277,16.3872095 8.48255835,18.3697677 12.0127909,18.3697677 C15.5430235,18.3697677 18.395349,16.3872095 18.395349,13.9313956 C18.395349,13.7651165 18.3825583,13.6116281 18.356977,13.4581397 C18.8813956,13.2023258 19.2779072,12.6395351 19.2779072,11.9872095 C19.2779072,11.1046514 18.5616281,10.3883723 17.67907,10.3883723 C17.2441863,10.3883723 16.8604653,10.5546514 16.57907,10.8360467 C15.4918607,10.0558142 13.9825583,9.54418626 12.3197677,9.48023277 L13.0488374,6.06511649 L15.4151165,6.5639537 C15.4406979,7.16511649 15.9395351,7.651163 16.5534886,7.651163 C17.1802328,7.651163 17.6918607,7.13953509 17.6918607,6.51279091 C17.6918607,5.88604672 17.1802328,5.37441881 16.5534886,5.37441881 Z M9.72325602,15.7093025 C10.2093025,16.195349 11.2581397,16.3744188 12.0127909,16.3744188 C12.7674421,16.3744188 13.8034886,16.195349 14.3023258,15.7093025 C14.4174421,15.5941863 14.6093025,15.5941863 14.7244188,15.7093025 C14.8139537,15.8372095 14.8139537,16.0162793 14.6988374,16.1313956 C13.9186049,16.9116281 12.4348839,16.9627909 12.0000002,16.9627909 C11.5651165,16.9627909 10.0686049,16.8988374 9.301163,16.1313956 C9.18604672,16.0162793 9.18604672,15.8244188 9.301163,15.7093025 C9.41627928,15.5941863 9.60813974,15.5941863 9.72325602,15.7093025 Z M9.48023277,11.9872095 C10.1069769,11.9872095 10.6186049,12.4988374 10.6186049,13.1255816 C10.6186049,13.7523258 10.1069769,14.2639537 9.48023277,14.2639537 C8.85348858,14.2639537 8.34186067,13.7523258 8.34186067,13.1255816 C8.34186067,12.4988374 8.85348858,11.9872095 9.48023277,11.9872095 Z M14.4941863,11.9872095 C15.1209304,11.9872095 15.6325583,12.4988374 15.6325583,13.1255816 C15.6325583,13.7523258 15.1209304,14.2639537 14.4941863,14.2639537 C13.8674421,14.2639537 13.3558142,13.7523258 13.3558142,13.1255816 C13.3558142,12.4988374 13.8674421,11.9872095 14.4941863,11.9872095 Z" id="Combined-Shape" fill="#212121" fill-rule="nonzero"></path>
</g>
</g>
</svg>
</a>
</li>
<li>
<a class="social-media-sharer" href="https://toot.kytta.dev/?text=Some%20article%20title%20https%3A%2F%2Fexample.com%2Fsome-article-url" target="_blank" rel="noopener noreferrer" aria-label="Share this page on Mastodon">
<svg width="24px" height="24px">
<g id="mastodon" stroke="none" stroke-width="1" fill="none" fill-rule="evenodd">
<g>
<rect id="Rectangle" x="0" y="0" width="24" height="24"></rect>
<path d='M11.5412949,0 C14.6936837,0.0250851513 17.7279692,0.357269448 19.4950021,1.1471931 C19.4950021,1.1471931 22.9995752,2.67324958 22.9995752,7.87958222 L23,8.27840764 C22.9948717,9.32900538 22.9399626,12.2394095 22.5108214,14.3877566 C22.1727192,16.0807457 19.4826471,17.9335557 16.3930961,18.2926354 C14.7820287,18.4797396 13.1958041,18.6517151 11.5043627,18.5762011 C8.73816776,18.4528442 6.55544838,17.9335557 6.55544838,17.9335557 C6.55544838,18.1956567 6.57205459,18.4452152 6.605267,18.6786105 C6.96489093,21.335697 9.312211,21.4948713 11.5357152,21.5690923 C13.7799439,21.6438306 15.7782681,21.0305374 15.7782681,21.0305374 L15.8704657,23.0052819 C15.8704657,23.0052819 14.3007146,23.8257215 11.5043627,23.9766203 C9.96237703,24.0591169 8.04774824,23.9388633 5.8177344,23.3643616 C0.981210876,22.1183796 0.149439349,17.1004442 0.022169409,12.0089343 C-0.0166226796,10.4972307 0.00729025582,9.07177343 0.00729025582,7.87958222 C0.00729025582,2.67324958 3.51199627,1.1471931 3.51199627,1.1471931 C5.27916201,0.357269448 8.31145476,0.0250851513 11.4638436,0 L11.5412949,0 Z M15.8599153,5 C14.461897,5 13.4033065,5.46697151 12.7035195,6.40103745 L12.0229667,7.39238391 L11.3425554,6.40103745 C10.642627,5.46697151 9.58403654,5 8.18615964,5 C6.97793807,5 6.00462882,5.36912752 5.26142334,6.08919046 C4.54070495,6.8092534 4.18190148,7.78253068 4.18190148,9.00730143 L4.18190148,15 L6.91358821,15 L6.91358821,9.1834452 C6.91358821,7.95732233 7.50716259,7.33498047 8.69445278,7.33498047 C10.0071899,7.33498047 10.6652556,8.0732355 10.6652556,9.5330285 L10.6652556,12.7167687 L13.3808194,12.7167687 L13.3808194,9.5330285 C13.3808194,8.0732355 14.0387436,7.33498047 15.3514807,7.33498047 C16.5387709,7.33498047 17.1323453,7.95732233 17.1323453,9.1834452 L17.1323453,15 L19.864032,15 L19.864032,9.00730143 C19.864032,7.78253068 19.5052285,6.8092534 18.7846516,6.08919046 C18.0413047,5.36912752 17.0679954,5 15.8599153,5 Z' id='Combined-Shape' fill='#212121' fill-rule='nonzero'></path>
</g>
</g>
</svg>
</a>
</li>
</ul>
Social Media Sharers Journal Comment
HTML
<div class="social-media-sharers--wrapper">
<div data-hypothesis-trigger>
<span class="side-section-wrapper__link" data-behaviour="HypothesisTrigger"><span class="svg-background-image comment"></span><span class="visuallyhidden">Open annotations (there are currently <span data-hypothesis-annotation-count>0</span> annotations on this page). </span></span>
</div>
<ul class="social-media-sharers">
<li>
<a class="social-media-sharer email" href="mailto:?subject=Some%20article%20title&amp;body=https%3A%2F%2Fexample.com%2Fsome-article-url" target="_blank" rel="noopener noreferrer" aria-label="Share by Email">
<svg width="24px" height="24px" viewBox="0 0 24 24" version="1.1" xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink">
<g id="email" stroke="none" stroke-width="1" fill="none" fill-rule="evenodd">
<rect id="clear-button-bg" x="0" y="0" width="24" height="24"></rect>
<path d="M20,4 L4,4 C2.9,4 2.01,4.9 2.01,6 L2,18 C2,19.1 2.9,20 4,20 L20,20 C21.1,20 22,19.1 22,18 L22,6 C22,4.9 21.1,4 20,4 Z M20,8 L12,13 L4,8 L4,6 L12,11 L20,6 L20,8 Z" id="Shape" fill="#000000" fill-rule="nonzero"></path>
</g>
</svg>
</a>
</li>
<li>
<a class="social-media-sharer" href="https://twitter.com/intent/tweet/?text=Some%20article%20title&amp;url=https%3A%2F%2Fexample.com%2Fsome-article-url" target="_blank" rel="noopener noreferrer" aria-label="Tweet a link to this page">
<svg width="24px" height="24px" viewBox="0 0 24 24" version="1.1" xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink">
<g id="x-24-articles" stroke="none" stroke-width="1" fill="none" fill-rule="evenodd">
<rect id="Rectangle" x="0" y="0" width="24" height="24"></rect>
<path d="M13.6604555,11.4785798 L20.954365,3 L19.225942,3 L12.8926412,10.3618317 L7.83425247,3 L2,3 L9.64927632,14.1323934 L2,23.023486 L3.72852106,23.023486 L10.4166498,15.2491415 L15.7586789,23.023486 L21.5929313,23.023486 L13.660031,11.4785798 L13.6604555,11.4785798 Z M11.293009,14.2304723 L10.5179779,13.1219369 L4.35133136,4.30120576 L7.00623887,4.30120576 L11.9827944,11.4198173 L12.7578255,12.5283527 L19.2267583,21.7814574 L16.5718508,21.7814574 L11.293009,14.2308968 L11.293009,14.2304723 Z" id="Shape" fill="#212121" fill-rule="nonzero"></path>
</g>
</svg>
</a>
</li>
<li>
<a class="social-media-sharer" href="https://facebook.com/sharer/sharer.php?u=https%3A%2F%2Fexample.com%2Fsome-article-url" target="_blank" rel="noopener noreferrer" aria-label="Share on Facebook">
<svg width="24px" height="24px" viewBox="0 0 24 24" version="1.1" xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink">
<g id="facebook" stroke="none" stroke-width="1" fill="none" fill-rule="evenodd">
<g>
<rect id="clear-button-bg" x="0" y="0" width="24" height="24"></rect>
<path d="M11.8969072,2 C6.43099227,2 2,6.41067993 2,11.8515383 C2,16.7687258 5.61915593,20.844338 10.3505155,21.5833958 L10.3505155,14.6992486 L7.83762887,14.6992486 L7.83762887,11.8515383 L10.3505155,11.8515383 L10.3505155,9.68112126 C10.3505155,7.21207948 11.8280541,5.84825714 14.0887242,5.84825714 C15.1715271,5.84825714 16.3041237,6.04067 16.3041237,6.04067 L16.3041237,8.465072 L15.0561469,8.465072 C13.8267075,8.465072 13.443299,9.22446783 13.443299,10.0035475 L13.443299,11.8515383 L16.1881443,11.8515383 L15.7493557,14.6992486 L13.443299,14.6992486 L13.443299,21.5833958 C18.1746585,20.844338 21.7938144,16.7687258 21.7938144,11.8515383 C21.7938144,6.41067993 17.3628222,2 11.8969072,2 Z" id="Fill-1" fill="#212121" fill-rule="nonzero"></path>
</g>
</g>
</svg>
</a>
</li>
<li>
<a class="social-media-sharer" href="https://www.linkedin.com/shareArticle?url=https%3A%2F%2Fexample.com%2Fsome-article-url" target="_self" aria-label="Share this page to LinkedIn (opens up email program, if configured on this system)">
<svg width="24px" height="24px" viewBox="0 0 24 24" version="1.1" xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink">
<g id="linkedin" stroke="none" stroke-width="1" fill="none" fill-rule="evenodd">
<g>
<rect id="Rectangle" x="0" y="0" width="24" height="24"></rect>
<path d="M18.9751954,18.9751435 L16.0232549,18.9751435 L16.0232549,14.3522816 C16.0232549,13.2499163 16.0035753,11.8308324 14.4879383,11.8308324 C12.9504693,11.8308324 12.7152366,13.0319032 12.7152366,14.2720257 L12.7152366,18.974836 L9.76329605,18.974836 L9.76329605,9.46835755 L12.5971589,9.46835755 L12.5971589,10.7675189 L12.6368256,10.7675189 C13.2146663,9.77952684 14.2890899,9.18943873 15.4328668,9.23189481 C18.4247815,9.23189481 18.9764254,11.1998552 18.9764254,13.7600486 L18.9751954,18.9751435 Z M6.43252317,8.16888879 C5.48643292,8.16905854 4.71933758,7.40223852 4.7191677,6.45614827 C4.71899794,5.51005802 5.48581795,4.74296266 6.4319082,4.74279278 C7.37799845,4.74262301 8.14509382,5.50944301 8.14526371,6.45553326 C8.14534522,6.90986184 7.96494211,7.34561469 7.64374096,7.66693118 C7.32253981,7.98824766 6.88685175,8.16880719 6.43252317,8.16888879 M7.90849343,18.9751435 L4.95347798,18.9751435 L4.95347798,9.46835755 L7.90849343,9.46835755 L7.90849343,18.9751435 Z M20.4468607,2.00148552 L3.47012787,2.00148552 C2.66777241,1.99243094 2.00979231,2.63513479 2,3.43748159 L2,20.4846305 C2.00945705,21.2873639 2.6673842,21.9307041 3.47012787,21.9223954 L20.4468607,21.9223954 C21.2511925,21.9322515 21.9116962,21.2889485 21.922831,20.4846305 L21.922831,3.43625161 C21.9113617,2.63231978 21.2508045,1.98965267 20.4468607,2" id="Path_2520" fill="#212121" fill-rule="nonzero"></path>
</g>
</g>
</svg>
</a>
</li>
<li>
<a class="social-media-sharer" href="https://reddit.com/submit/?url=https%3A%2F%2Fexample.com%2Fsome-article-url" target="_blank" rel="noopener noreferrer" aria-label="Share this page on Reddit">
<svg width="24px" height="24px" viewBox="0 0 24 24" version="1.1" xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink">
<g id="reddit" stroke="none" stroke-width="1" fill="none" fill-rule="evenodd">
<g>
<rect id="Rectangle" x="0" y="0" width="24" height="24"></rect>
<path d="M11.9872095,1.051163 C18.0270212,1.051163 22.923256,5.94739779 22.923256,11.9872095 C22.923256,18.0270212 18.0270212,22.923256 11.9872095,22.923256 C5.94739779,22.923256 1.051163,18.0270212 1.051163,11.9872095 C1.051163,5.94739779 5.94739779,1.051163 11.9872095,1.051163 Z M16.5534886,5.37441881 C16.1058142,5.37441881 15.7220932,5.63023277 15.5430235,6.0139537 L12.895349,5.451163 C12.8186049,5.4383723 12.7418607,5.451163 12.6779072,5.48953509 C12.6139537,5.52790719 12.5755816,5.59186067 12.5500002,5.66860486 L11.7441863,9.48023277 C10.0430235,9.53139556 8.52093044,10.0302328 7.42093044,10.8360467 C7.13953509,10.5674421 6.74302347,10.3883723 6.32093044,10.3883723 C5.4383723,10.3883723 4.72209323,11.1046514 4.72209323,11.9872095 C4.72209323,12.6395351 5.10581416,13.1895351 5.66860486,13.445349 C5.64302347,13.5988374 5.63023277,13.7651165 5.63023277,13.9313956 C5.63023277,16.3872095 8.48255835,18.3697677 12.0127909,18.3697677 C15.5430235,18.3697677 18.395349,16.3872095 18.395349,13.9313956 C18.395349,13.7651165 18.3825583,13.6116281 18.356977,13.4581397 C18.8813956,13.2023258 19.2779072,12.6395351 19.2779072,11.9872095 C19.2779072,11.1046514 18.5616281,10.3883723 17.67907,10.3883723 C17.2441863,10.3883723 16.8604653,10.5546514 16.57907,10.8360467 C15.4918607,10.0558142 13.9825583,9.54418626 12.3197677,9.48023277 L13.0488374,6.06511649 L15.4151165,6.5639537 C15.4406979,7.16511649 15.9395351,7.651163 16.5534886,7.651163 C17.1802328,7.651163 17.6918607,7.13953509 17.6918607,6.51279091 C17.6918607,5.88604672 17.1802328,5.37441881 16.5534886,5.37441881 Z M9.72325602,15.7093025 C10.2093025,16.195349 11.2581397,16.3744188 12.0127909,16.3744188 C12.7674421,16.3744188 13.8034886,16.195349 14.3023258,15.7093025 C14.4174421,15.5941863 14.6093025,15.5941863 14.7244188,15.7093025 C14.8139537,15.8372095 14.8139537,16.0162793 14.6988374,16.1313956 C13.9186049,16.9116281 12.4348839,16.9627909 12.0000002,16.9627909 C11.5651165,16.9627909 10.0686049,16.8988374 9.301163,16.1313956 C9.18604672,16.0162793 9.18604672,15.8244188 9.301163,15.7093025 C9.41627928,15.5941863 9.60813974,15.5941863 9.72325602,15.7093025 Z M9.48023277,11.9872095 C10.1069769,11.9872095 10.6186049,12.4988374 10.6186049,13.1255816 C10.6186049,13.7523258 10.1069769,14.2639537 9.48023277,14.2639537 C8.85348858,14.2639537 8.34186067,13.7523258 8.34186067,13.1255816 C8.34186067,12.4988374 8.85348858,11.9872095 9.48023277,11.9872095 Z M14.4941863,11.9872095 C15.1209304,11.9872095 15.6325583,12.4988374 15.6325583,13.1255816 C15.6325583,13.7523258 15.1209304,14.2639537 14.4941863,14.2639537 C13.8674421,14.2639537 13.3558142,13.7523258 13.3558142,13.1255816 C13.3558142,12.4988374 13.8674421,11.9872095 14.4941863,11.9872095 Z" id="Combined-Shape" fill="#212121" fill-rule="nonzero"></path>
</g>
</g>
</svg>
</a>
</li>
<li>
<a class="social-media-sharer" href="https://toot.kytta.dev/?text=Some%20article%20title%20https%3A%2F%2Fexample.com%2Fsome-article-url" target="_blank" rel="noopener noreferrer" aria-label="Share this page on Mastodon">
<svg width="24px" height="24px">
<g id="mastodon" stroke="none" stroke-width="1" fill="none" fill-rule="evenodd">
<g>
<rect id="Rectangle" x="0" y="0" width="24" height="24"></rect>
<path d='M11.5412949,0 C14.6936837,0.0250851513 17.7279692,0.357269448 19.4950021,1.1471931 C19.4950021,1.1471931 22.9995752,2.67324958 22.9995752,7.87958222 L23,8.27840764 C22.9948717,9.32900538 22.9399626,12.2394095 22.5108214,14.3877566 C22.1727192,16.0807457 19.4826471,17.9335557 16.3930961,18.2926354 C14.7820287,18.4797396 13.1958041,18.6517151 11.5043627,18.5762011 C8.73816776,18.4528442 6.55544838,17.9335557 6.55544838,17.9335557 C6.55544838,18.1956567 6.57205459,18.4452152 6.605267,18.6786105 C6.96489093,21.335697 9.312211,21.4948713 11.5357152,21.5690923 C13.7799439,21.6438306 15.7782681,21.0305374 15.7782681,21.0305374 L15.8704657,23.0052819 C15.8704657,23.0052819 14.3007146,23.8257215 11.5043627,23.9766203 C9.96237703,24.0591169 8.04774824,23.9388633 5.8177344,23.3643616 C0.981210876,22.1183796 0.149439349,17.1004442 0.022169409,12.0089343 C-0.0166226796,10.4972307 0.00729025582,9.07177343 0.00729025582,7.87958222 C0.00729025582,2.67324958 3.51199627,1.1471931 3.51199627,1.1471931 C5.27916201,0.357269448 8.31145476,0.0250851513 11.4638436,0 L11.5412949,0 Z M15.8599153,5 C14.461897,5 13.4033065,5.46697151 12.7035195,6.40103745 L12.0229667,7.39238391 L11.3425554,6.40103745 C10.642627,5.46697151 9.58403654,5 8.18615964,5 C6.97793807,5 6.00462882,5.36912752 5.26142334,6.08919046 C4.54070495,6.8092534 4.18190148,7.78253068 4.18190148,9.00730143 L4.18190148,15 L6.91358821,15 L6.91358821,9.1834452 C6.91358821,7.95732233 7.50716259,7.33498047 8.69445278,7.33498047 C10.0071899,7.33498047 10.6652556,8.0732355 10.6652556,9.5330285 L10.6652556,12.7167687 L13.3808194,12.7167687 L13.3808194,9.5330285 C13.3808194,8.0732355 14.0387436,7.33498047 15.3514807,7.33498047 C16.5387709,7.33498047 17.1323453,7.95732233 17.1323453,9.1834452 L17.1323453,15 L19.864032,15 L19.864032,9.00730143 C19.864032,7.78253068 19.5052285,6.8092534 18.7846516,6.08919046 C18.0413047,5.36912752 17.0679954,5 15.8599153,5 Z' id='Combined-Shape' fill='#212121' fill-rule='nonzero'></path>
</g>
</g>
</svg>
</a>
</li>
</ul>
</div>
Social Media Sharers
HTML
<div class="social-media-sharers">
<a class="social-media-sharer" href="https://facebook.com/sharer/sharer.php?u=https%3A%2F%2Fexample.com%2Fsome-article-url" target="_blank" rel="noopener noreferrer" aria-label="Share on Facebook">
<div class="social-media-sharer__icon_wrapper social-media-sharer__icon_wrapper--facebook social-media-sharer__icon_wrapper--small"><div aria-hidden="true" class="social-media-sharer__icon social-media-sharer__icon--solid">
<svg xmlns="http://www.w3.org/2000/svg" viewBox="0 0 24 24"><path d="M18.77 7.46H14.5v-1.9c0-.9.6-1.1 1-1.1h3V.5h-4.33C10.24.5 9.5 3.44 9.5 5.32v2.15h-3v4h3v12h5v-12h3.85l.42-4z"/></svg>
</div>
</div>
</a>
<a class="social-media-sharer" href="https://twitter.com/intent/tweet/?text=Some%20article%20title&amp;url=https%3A%2F%2Fexample.com%2Fsome-article-url" target="_blank" rel="noopener noreferrer" aria-label="Tfweet a link to this page">
<div class="social-media-sharer__icon_wrapper social-media-sharer__icon_wrapper--twitter social-media-sharer__icon_wrapper--small">
<div aria-hidden="true" class="social-media-sharer__icon social-media-sharer__icon--solid">
<svg xmlns="http://www.w3.org/2000/svg" viewBox="0 0 24 24">
<path d="M13.6604555,11.4785798 L20.954365,3 L19.225942,3 L12.8926412,10.3618317 L7.83425247,3 L2,3 L9.64927632,14.1323934 L2,23.023486 L3.72852106,23.023486 L10.4166498,15.2491415 L15.7586789,23.023486 L21.5929313,23.023486 L13.660031,11.4785798 L13.6604555,11.4785798 Z M11.293009,14.2304723 L10.5179779,13.1219369 L4.35133136,4.30120576 L7.00623887,4.30120576 L11.9827944,11.4198173 L12.7578255,12.5283527 L19.2267583,21.7814574 L16.5718508,21.7814574 L11.293009,14.2308968 L11.293009,14.2304723 Z" />
</svg>
</div>
</div>
</a>
<a class="social-media-sharer" href="mailto:?subject=Some%20article%20title&amp;body=https%3A%2F%2Fexample.com%2Fsome-article-url" target="_self" aria-label="Email a link to this page (opens up email program, if configured on this system)">
<div class="social-media-sharer__icon_wrapper social-media-sharer__icon_wrapper--email social-media-sharer__icon_wrapper--small"><div aria-hidden="true" class="social-media-sharer__icon social-media-sharer__icon--solid">
<svg xmlns="http://www.w3.org/2000/svg" viewBox="0 0 24 24"><path d="M22 4H2C.9 4 0 4.9 0 6v12c0 1.1.9 2 2 2h20c1.1 0 2-.9 2-2V6c0-1.1-.9-2-2-2zM7.25 14.43l-3.5 2c-.08.05-.17.07-.25.07-.17 0-.34-.1-.43-.25-.14-.24-.06-.55.18-.68l3.5-2c.24-.14.55-.06.68.18.14.24.06.55-.18.68zm4.75.07c-.1 0-.2-.03-.27-.08l-8.5-5.5c-.23-.15-.3-.46-.15-.7.15-.22.46-.3.7-.14L12 13.4l8.23-5.32c.23-.15.54-.08.7.15.14.23.07.54-.16.7l-8.5 5.5c-.08.04-.17.07-.27.07zm8.93 1.75c-.1.16-.26.25-.43.25-.08 0-.17-.02-.25-.07l-3.5-2c-.24-.13-.32-.44-.18-.68s.44-.32.68-.18l3.5 2c.24.13.32.44.18.68z"/></svg>
</div>
</div>
</a>
<a class="social-media-sharer" href="https://reddit.com/submit/?url=https%3A%2F%2Fexample.com%2Fsome-article-url" target="_blank" rel="noopener noreferrer" aria-label="Share this page on Reddit">
<div class="social-media-sharer__icon_wrapper social-media-sharer__icon_wrapper--reddit social-media-sharer__icon_wrapper--small"><div aria-hidden="true" class="social-media-sharer__icon social-media-sharer__icon--solid">
<svg xmlns="http://www.w3.org/2000/svg" viewBox="0 0 24 24"><path d="M24 11.5c0-1.65-1.35-3-3-3-.96 0-1.86.48-2.42 1.24-1.64-1-3.75-1.64-6.07-1.72.08-1.1.4-3.05 1.52-3.7.72-.4 1.73-.24 3 .5C17.2 6.3 18.46 7.5 20 7.5c1.65 0 3-1.35 3-3s-1.35-3-3-3c-1.38 0-2.54.94-2.88 2.22-1.43-.72-2.64-.8-3.6-.25-1.64.94-1.95 3.47-2 4.55-2.33.08-4.45.7-6.1 1.72C4.86 8.98 3.96 8.5 3 8.5c-1.65 0-3 1.35-3 3 0 1.32.84 2.44 2.05 2.84-.03.22-.05.44-.05.66 0 3.86 4.5 7 10 7s10-3.14 10-7c0-.22-.02-.44-.05-.66 1.2-.4 2.05-1.54 2.05-2.84zM2.3 13.37C1.5 13.07 1 12.35 1 11.5c0-1.1.9-2 2-2 .64 0 1.22.32 1.6.82-1.1.85-1.92 1.9-2.3 3.05zm3.7.13c0-1.1.9-2 2-2s2 .9 2 2-.9 2-2 2-2-.9-2-2zm9.8 4.8c-1.08.63-2.42.96-3.8.96-1.4 0-2.74-.34-3.8-.95-.24-.13-.32-.44-.2-.68.15-.24.46-.32.7-.18 1.83 1.06 4.76 1.06 6.6 0 .23-.13.53-.05.67.2.14.23.06.54-.18.67zm.2-2.8c-1.1 0-2-.9-2-2s.9-2 2-2 2 .9 2 2-.9 2-2 2zm5.7-2.13c-.38-1.16-1.2-2.2-2.3-3.05.38-.5.97-.82 1.6-.82 1.1 0 2 .9 2 2 0 .84-.53 1.57-1.3 1.87z"/></svg>
</div>
</div>
</a>
</div>
Speech Bubble
HTML
<button class="speech-bubble"
data-behaviour="SpeechBubble"
aria-live="polite">
<span class="speech-bubble__inner">8</span>
</button>
Speech Bubble Has Placeholder
HTML
<button class="speech-bubble speech-bubble--has-placeholder"
data-behaviour="SpeechBubble"
aria-live="polite">
<span class="speech-bubble__inner">+</span>
</button>
Speech Bubble Share Your Feedback
HTML
<button class="speech-bubble speech-bubble--wrapped "
data-behaviour="SpeechBubble"
aria-live="polite">
<span class="speech-bubble--wrapped__prefix">Add a comment</span>
<span class="speech-bubble__inner">999</span>
</button>
Speech Bubble Small Is Loading
HTML
<button class="speech-bubble speech-bubble--small speech-bubble--loading"
data-behaviour="SpeechBubble"
aria-live="polite">
<span class="speech-bubble__inner"></span>
</button>
Speech Bubble Small
HTML
<button class="speech-bubble speech-bubble--small "
data-behaviour="SpeechBubble"
aria-live="polite">
<span class="speech-bubble__inner">340</span>
</button>
Speech Bubble Wrapped Has Placeholder
HTML
<button class="speech-bubble speech-bubble--has-placeholder speech-bubble--wrapped "
data-behaviour="SpeechBubble"
aria-live="polite">
<span class="speech-bubble--wrapped__prefix">Add a comment</span>
<span class="speech-bubble__inner">+</span>
</button>
Table
| F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | Partial η2 | Original effect size f | Replication total sample size | Detectable effect size f |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F(24,39) = 0.8678 (interaction) | 0.348120 | 0.7307699 | 169* | 0.3895070 | ||||||||
| F(2,39) = 0.8075 (treatments) | 0.039766 | 0.2035014 | 169 | 0.2415459 | ||||||||
| F(12,39) = 187.6811 (hematology parameters) | 0.982978 | 7.599178 | 169* | 0.3331365 |
| F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | F(Dfn, Dfd) | Partial η2 | Original effect size f | Replication total sample size | Detectable effect size f |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F(24,39) = 0.8678 (interaction) | 0.348120 | 0.7307699 | 169* | 0.3895070 | ||||||||
| F(2,39) = 0.8075 (treatments) | 0.039766 | 0.2035014 | 169* | 0.2415459 | ||||||||
| F(12,39) = 187.6811 (hematology parameters) | 0.982978 | 7.599178 | 169* | 0.3331365 |
-
*
Footnote 1.
-
The values in parenthesis refer to the highest resolution shell.
*Rmerge=∑hkl∑i|I(hkl;i)−<I(hkl)>|∑hkl∑i(hkl;i) where I(hkl;i) is the intensity of an individual measurement of a reflection and <I(hkl)> is the average intensity of that reflection.
HTML
<div class="table">
<div class="table__table">
<table><thead><tr><th>F(Dfn, Dfd)</th><th>F(Dfn, Dfd)</th><th>F(Dfn, Dfd)</th><th>F(Dfn, Dfd)</th><th>F(Dfn, Dfd)</th><th>F(Dfn, Dfd)</th><th>F(Dfn, Dfd)</th><th>F(Dfn, Dfd)</th><th>F(Dfn, Dfd)</th><th>Partial η<sup>2</sup></th><th>Original effect size <em>f</em></th><th>Replication total sample size</th><th>Detectable effect size <em>f</em></th></tr></thead><tbody><tr><td>F(24,39) = 0.8678 (interaction)</td><td>0.348120</td><td>0.7307699</td><td>169<a href="#tblfn1">*</a></td><td>0.3895070</td></tr><tr><td>F(2,39) = 0.8075 (treatments)</td><td>0.039766</td><td>0.2035014</td><td>169</td><td>0.2415459</td></tr><tr><td>F(12,39) = 187.6811 (hematology parameters)</td><td>0.982978</td><td>7.599178</td><td>169<a href="#tblfn1">*</a></td><td>0.3331365</td></tr></tbody></table>
</div>
<div class="table__table">
<table><thead><tr><th>F(Dfn, Dfd)</th><th>F(Dfn, Dfd)</th><th>F(Dfn, Dfd)</th><th>F(Dfn, Dfd)</th><th>F(Dfn, Dfd)</th><th>F(Dfn, Dfd)</th><th>F(Dfn, Dfd)</th><th>F(Dfn, Dfd)</th><th>F(Dfn, Dfd)</th><th>Partial η<sup>2</sup></th><th>Original effect size <em>f</em></th><th>Replication total sample size</th><th>Detectable effect size <em>f</em></th></tr></thead><tbody><tr><td>F(24,39) = 0.8678 (interaction)</td><td>0.348120</td><td>0.7307699</td><td>169<a href="#tblfn1">*</a></td><td>0.3895070</td></tr><tr><td>F(2,39) = 0.8075 (treatments)</td><td>0.039766</td><td>0.2035014</td><td>169<a href="#tblfn1">*</a></td><td>0.2415459</td></tr><tr><td>F(12,39) = 187.6811 (hematology parameters)</td><td>0.982978</td><td>7.599178</td><td>169<a href="#tblfn1">*</a></td><td>0.3331365</td></tr></tbody></table>
</div>
<ol class="list">
<li class="table-footnote" id="tblfn1">
<div class="table-footnote__label">*</div>
<div class="table-footnote__text"><p>Footnote 1.</p></div>
</li>
<li class="table-footnote" >
<div class="table-footnote__text"><p>The values in parenthesis refer to the highest resolution shell.</p><p><sup>*</sup><span class="MathJax_Preview" style="display: none;"></span><span id="MathJax-Element-1-Frame" class="mjx-chtml MathJax_CHTML" tabindex="0" data-mathml="<math xmlns="http://www.w3.org/1998/Math/MathML" id="inf1"><mstyle displaystyle="true" scriptlevel="0"><mrow><msub><mi>R</mi><mrow><mi>m</mi><mi>e</mi><mi>r</mi><mi>g</mi><mi>e</mi></mrow></msub><mo>=</mo><mfrac><mrow><msub><mo movablelimits="false">&#x2211;</mo><mrow><mi>h</mi><mi>k</mi><mi>l</mi></mrow></msub><msub><mo movablelimits="false">&#x2211;</mo><mrow><mi>i</mi></mrow></msub><mrow><mo stretchy="false">|</mo></mrow><mi>I</mi><mo stretchy="false">(</mo><mi>h</mi><mi>k</mi><mi>l</mi><mo>;</mo><mi>i</mi><mo stretchy="false">)</mo><mo>&#x2212;</mo><mo>&lt;</mo><mi>I</mi><mo stretchy="false">(</mo><mi>h</mi><mi>k</mi><mi>l</mi><mo stretchy="false">)</mo><mo>&gt;</mo><mrow><mo stretchy="false">|</mo></mrow></mrow><mrow><msub><mo movablelimits="false">&#x2211;</mo><mrow><mi>h</mi><mi>k</mi><mi>l</mi></mrow></msub><msub><mo movablelimits="false">&#x2211;</mo><mrow><mi>i</mi></mrow></msub><mo stretchy="false">(</mo><mi>h</mi><mi>k</mi><mi>l</mi><mo>;</mo><mi>i</mi><mo stretchy="false">)</mo></mrow></mfrac></mrow></mstyle></math>" role="presentation" style="font-size: 107%; position: relative;"><span id="inf1" class="mjx-math" aria-hidden="true"><span id="MJXc-Node-2" class="mjx-mrow"><span id="MJXc-Node-3" class="mjx-mstyle"><span id="MJXc-Node-4" class="mjx-mrow"><span id="MJXc-Node-5" class="mjx-mrow"><span id="MJXc-Node-6" class="mjx-msub"><span class="mjx-base"><span id="MJXc-Node-7" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.435em; padding-bottom: 0.291em;">R</span></span></span><span class="mjx-sub" style="font-size: 70.7%; vertical-align: -0.212em; padding-right: 0.071em;"><span id="MJXc-Node-8" class="mjx-mrow" style=""><span id="MJXc-Node-9" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.219em; padding-bottom: 0.291em;">m</span></span><span id="MJXc-Node-10" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.219em; padding-bottom: 0.291em;">e</span></span><span id="MJXc-Node-11" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.219em; padding-bottom: 0.291em;">r</span></span><span id="MJXc-Node-12" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.219em; padding-bottom: 0.507em; padding-right: 0.003em;">g</span></span><span id="MJXc-Node-13" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.219em; padding-bottom: 0.291em;">e</span></span></span></span></span><span id="MJXc-Node-14" class="mjx-mo MJXc-space3"><span class="mjx-char MJXc-TeX-main-R" style="padding-top: 0.075em; padding-bottom: 0.291em;">=</span></span><span id="MJXc-Node-15" class="mjx-mfrac MJXc-space3"><span class="mjx-box MJXc-stacked" style="width: 13.813em; padding: 0px 0.12em;"><span class="mjx-numerator" style="width: 13.813em; top: -1.645em;"><span id="MJXc-Node-16" class="mjx-mrow"><span id="MJXc-Node-17" class="mjx-msub"><span class="mjx-base"><span id="MJXc-Node-18" class="mjx-mo"><span class="mjx-char MJXc-TeX-size1-R" style="padding-top: 0.507em; padding-bottom: 0.507em;">∑</span></span></span><span class="mjx-sub" style="font-size: 70.7%; vertical-align: -0.439em; padding-right: 0.071em;"><span id="MJXc-Node-19" class="mjx-mrow" style=""><span id="MJXc-Node-20" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.507em; padding-bottom: 0.291em;">h</span></span><span id="MJXc-Node-21" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.507em; padding-bottom: 0.291em;">k</span></span><span id="MJXc-Node-22" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.507em; padding-bottom: 0.291em;">l</span></span></span></span></span><span id="MJXc-Node-23" class="mjx-msub MJXc-space1"><span class="mjx-base"><span id="MJXc-Node-24" class="mjx-mo"><span class="mjx-char MJXc-TeX-size1-R" style="padding-top: 0.507em; padding-bottom: 0.507em;">∑</span></span></span><span class="mjx-sub" style="font-size: 70.7%; vertical-align: -0.439em; padding-right: 0.071em;"><span id="MJXc-Node-25" class="mjx-mrow" style=""><span id="MJXc-Node-26" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.435em; padding-bottom: 0.291em;">i</span></span></span></span></span><span id="MJXc-Node-27" class="mjx-mrow MJXc-space1"><span id="MJXc-Node-28" class="mjx-mo"><span class="mjx-char MJXc-TeX-main-R" style="padding-top: 0.435em; padding-bottom: 0.579em;">|</span></span></span><span id="MJXc-Node-29" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.435em; padding-bottom: 0.291em; padding-right: 0.064em;">I</span></span><span id="MJXc-Node-30" class="mjx-mo"><span class="mjx-char MJXc-TeX-main-R" style="padding-top: 0.435em; padding-bottom: 0.579em;">(</span></span><span id="MJXc-Node-31" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.507em; padding-bottom: 0.291em;">h</span></span><span id="MJXc-Node-32" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.507em; padding-bottom: 0.291em;">k</span></span><span id="MJXc-Node-33" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.507em; padding-bottom: 0.291em;">l</span></span><span id="MJXc-Node-34" class="mjx-mo"><span class="mjx-char MJXc-TeX-main-R" style="padding-top: 0.147em; padding-bottom: 0.507em;">;</span></span><span id="MJXc-Node-35" class="mjx-mi MJXc-space1"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.435em; padding-bottom: 0.291em;">i</span></span><span id="MJXc-Node-36" class="mjx-mo"><span class="mjx-char MJXc-TeX-main-R" style="padding-top: 0.435em; padding-bottom: 0.579em;">)</span></span><span id="MJXc-Node-37" class="mjx-mo"><span class="mjx-char MJXc-TeX-main-R" style="padding-top: 0.291em; padding-bottom: 0.435em;">−</span></span><span id="MJXc-Node-38" class="mjx-mo MJXc-space3"><span class="mjx-char MJXc-TeX-main-R" style="padding-top: 0.219em; padding-bottom: 0.363em;"><</span></span><span id="MJXc-Node-39" class="mjx-mi MJXc-space3"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.435em; padding-bottom: 0.291em; padding-right: 0.064em;">I</span></span><span id="MJXc-Node-40" class="mjx-mo"><span class="mjx-char MJXc-TeX-main-R" style="padding-top: 0.435em; padding-bottom: 0.579em;">(</span></span><span id="MJXc-Node-41" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.507em; padding-bottom: 0.291em;">h</span></span><span id="MJXc-Node-42" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.507em; padding-bottom: 0.291em;">k</span></span><span id="MJXc-Node-43" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.507em; padding-bottom: 0.291em;">l</span></span><span id="MJXc-Node-44" class="mjx-mo"><span class="mjx-char MJXc-TeX-main-R" style="padding-top: 0.435em; padding-bottom: 0.579em;">)</span></span><span id="MJXc-Node-45" class="mjx-mo MJXc-space3"><span class="mjx-char MJXc-TeX-main-R" style="padding-top: 0.219em; padding-bottom: 0.363em;">></span></span><span id="MJXc-Node-46" class="mjx-mrow"><span id="MJXc-Node-47" class="mjx-mo"><span class="mjx-char MJXc-TeX-main-R" style="padding-top: 0.435em; padding-bottom: 0.579em;">|</span></span></span></span></span><span class="mjx-denominator" style="width: 13.813em; bottom: -1.145em;"><span id="MJXc-Node-48" class="mjx-mrow"><span id="MJXc-Node-49" class="mjx-msub"><span class="mjx-base"><span id="MJXc-Node-50" class="mjx-mo"><span class="mjx-char MJXc-TeX-size1-R" style="padding-top: 0.507em; padding-bottom: 0.507em;">∑</span></span></span><span class="mjx-sub" style="font-size: 70.7%; vertical-align: -0.439em; padding-right: 0.071em;"><span id="MJXc-Node-51" class="mjx-mrow" style=""><span id="MJXc-Node-52" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.507em; padding-bottom: 0.291em;">h</span></span><span id="MJXc-Node-53" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.507em; padding-bottom: 0.291em;">k</span></span><span id="MJXc-Node-54" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.507em; padding-bottom: 0.291em;">l</span></span></span></span></span><span id="MJXc-Node-55" class="mjx-msub MJXc-space1"><span class="mjx-base"><span id="MJXc-Node-56" class="mjx-mo"><span class="mjx-char MJXc-TeX-size1-R" style="padding-top: 0.507em; padding-bottom: 0.507em;">∑</span></span></span><span class="mjx-sub" style="font-size: 70.7%; vertical-align: -0.439em; padding-right: 0.071em;"><span id="MJXc-Node-57" class="mjx-mrow" style=""><span id="MJXc-Node-58" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.435em; padding-bottom: 0.291em;">i</span></span></span></span></span><span id="MJXc-Node-59" class="mjx-mo"><span class="mjx-char MJXc-TeX-main-R" style="padding-top: 0.435em; padding-bottom: 0.579em;">(</span></span><span id="MJXc-Node-60" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.507em; padding-bottom: 0.291em;">h</span></span><span id="MJXc-Node-61" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.507em; padding-bottom: 0.291em;">k</span></span><span id="MJXc-Node-62" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.507em; padding-bottom: 0.291em;">l</span></span><span id="MJXc-Node-63" class="mjx-mo"><span class="mjx-char MJXc-TeX-main-R" style="padding-top: 0.147em; padding-bottom: 0.507em;">;</span></span><span id="MJXc-Node-64" class="mjx-mi MJXc-space1"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.435em; padding-bottom: 0.291em;">i</span></span><span id="MJXc-Node-65" class="mjx-mo"><span class="mjx-char MJXc-TeX-main-R" style="padding-top: 0.435em; padding-bottom: 0.579em;">)</span></span></span></span><span class="mjx-line" style="border-bottom: 1.3px solid; top: -0.295em; width: 13.813em;"></span></span><span class="mjx-vsize" style="height: 2.791em; vertical-align: -1.145em;"></span></span></span></span></span></span></span><span class="MJX_Assistive_MathML" role="presentation"><math xmlns="http://www.w3.org/1998/Math/MathML" id="inf1"><mstyle displaystyle="true" scriptlevel="0"><mrow><msub><mi>R</mi><mrow><mi>m</mi><mi>e</mi><mi>r</mi><mi>g</mi><mi>e</mi></mrow></msub><mo>=</mo><mfrac><mrow><msub><mo movablelimits="false">∑</mo><mrow><mi>h</mi><mi>k</mi><mi>l</mi></mrow></msub><msub><mo movablelimits="false">∑</mo><mrow><mi>i</mi></mrow></msub><mrow><mo stretchy="false">|</mo></mrow><mi>I</mi><mo stretchy="false">(</mo><mi>h</mi><mi>k</mi><mi>l</mi><mo>;</mo><mi>i</mi><mo stretchy="false">)</mo><mo>−</mo><mo><</mo><mi>I</mi><mo stretchy="false">(</mo><mi>h</mi><mi>k</mi><mi>l</mi><mo stretchy="false">)</mo><mo>></mo><mrow><mo stretchy="false">|</mo></mrow></mrow><mrow><msub><mo movablelimits="false">∑</mo><mrow><mi>h</mi><mi>k</mi><mi>l</mi></mrow></msub><msub><mo movablelimits="false">∑</mo><mrow><mi>i</mi></mrow></msub><mo stretchy="false">(</mo><mi>h</mi><mi>k</mi><mi>l</mi><mo>;</mo><mi>i</mi><mo stretchy="false">)</mo></mrow></mfrac></mrow></mstyle></math></span></span><script type="math/mml" id="MathJax-Element-1"><math id="inf1"><mstyle displaystyle="true" scriptlevel="0"><mrow><msub><mi>R</mi><mrow><mi>m</mi><mi>e</mi><mi>r</mi><mi>g</mi><mi>e</mi></mrow></msub><mo>=</mo><mfrac><mrow><msub><mo movablelimits="false">∑</mo><mrow><mi>h</mi><mi>k</mi><mi>l</mi></mrow></msub><msub><mo movablelimits="false">∑</mo><mrow><mi>i</mi></mrow></msub><mrow><mo stretchy="false">|</mo></mrow><mi>I</mi><mo stretchy="false">(</mo><mi>h</mi><mi>k</mi><mi>l</mi><mo>;</mo><mi>i</mi><mo stretchy="false">)</mo><mo>−</mo><mo><</mo><mi>I</mi><mo stretchy="false">(</mo><mi>h</mi><mi>k</mi><mi>l</mi><mo stretchy="false">)</mo><mo>></mo><mrow><mo stretchy="false">|</mo></mrow></mrow><mrow><msub><mo movablelimits="false">∑</mo><mrow><mi>h</mi><mi>k</mi><mi>l</mi></mrow></msub><msub><mo movablelimits="false">∑</mo><mrow><mi>i</mi></mrow></msub><mo stretchy="false">(</mo><mi>h</mi><mi>k</mi><mi>l</mi><mo>;</mo><mi>i</mi><mo stretchy="false">)</mo></mrow></mfrac></mrow></mstyle></math></script> where <span class="MathJax_Preview" style="display: none;"></span><span id="MathJax-Element-2-Frame" class="mjx-chtml MathJax_CHTML" tabindex="0" data-mathml="<math xmlns="http://www.w3.org/1998/Math/MathML" id="inf2"><mstyle displaystyle="true" scriptlevel="0"><mrow><mi>I</mi><mo stretchy="false">(</mo><mi>h</mi><mi>k</mi><mi>l</mi><mo>;</mo><mi>i</mi><mo stretchy="false">)</mo></mrow></mstyle></math>" role="presentation" style="font-size: 107%; position: relative;"><span id="inf2" class="mjx-math" aria-hidden="true"><span id="MJXc-Node-67" class="mjx-mrow"><span id="MJXc-Node-68" class="mjx-mstyle"><span id="MJXc-Node-69" class="mjx-mrow"><span id="MJXc-Node-70" class="mjx-mrow"><span id="MJXc-Node-71" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.435em; padding-bottom: 0.291em; padding-right: 0.064em;">I</span></span><span id="MJXc-Node-72" class="mjx-mo"><span class="mjx-char MJXc-TeX-main-R" style="padding-top: 0.435em; padding-bottom: 0.579em;">(</span></span><span id="MJXc-Node-73" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.507em; padding-bottom: 0.291em;">h</span></span><span id="MJXc-Node-74" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.507em; padding-bottom: 0.291em;">k</span></span><span id="MJXc-Node-75" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.507em; padding-bottom: 0.291em;">l</span></span><span id="MJXc-Node-76" class="mjx-mo"><span class="mjx-char MJXc-TeX-main-R" style="padding-top: 0.147em; padding-bottom: 0.507em;">;</span></span><span id="MJXc-Node-77" class="mjx-mi MJXc-space1"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.435em; padding-bottom: 0.291em;">i</span></span><span id="MJXc-Node-78" class="mjx-mo"><span class="mjx-char MJXc-TeX-main-R" style="padding-top: 0.435em; padding-bottom: 0.579em;">)</span></span></span></span></span></span></span><span class="MJX_Assistive_MathML" role="presentation"><math xmlns="http://www.w3.org/1998/Math/MathML" id="inf2"><mstyle displaystyle="true" scriptlevel="0"><mrow><mi>I</mi><mo stretchy="false">(</mo><mi>h</mi><mi>k</mi><mi>l</mi><mo>;</mo><mi>i</mi><mo stretchy="false">)</mo></mrow></mstyle></math></span></span><script type="math/mml" id="MathJax-Element-2"><math id="inf2"><mstyle displaystyle="true" scriptlevel="0"><mrow><mi>I</mi><mo stretchy="false">(</mo><mi>h</mi><mi>k</mi><mi>l</mi><mo>;</mo><mi>i</mi><mo stretchy="false">)</mo></mrow></mstyle></math></script> is the intensity of an individual measurement of a reflection and <span class="MathJax_Preview" style="display: none;"></span><span id="MathJax-Element-3-Frame" class="mjx-chtml MathJax_CHTML" tabindex="0" data-mathml="<math xmlns="http://www.w3.org/1998/Math/MathML" id="inf3"><mstyle displaystyle="true" scriptlevel="0"><mrow><mo>&lt;</mo><mi>I</mi><mo stretchy="false">(</mo><mi>h</mi><mi>k</mi><mi>l</mi><mo stretchy="false">)</mo><mo>&gt;</mo></mrow></mstyle></math>" role="presentation" style="font-size: 107%; position: relative;"><span id="inf3" class="mjx-math" aria-hidden="true"><span id="MJXc-Node-80" class="mjx-mrow"><span id="MJXc-Node-81" class="mjx-mstyle"><span id="MJXc-Node-82" class="mjx-mrow"><span id="MJXc-Node-83" class="mjx-mrow"><span id="MJXc-Node-84" class="mjx-mo"><span class="mjx-char MJXc-TeX-main-R" style="padding-top: 0.219em; padding-bottom: 0.363em;"><</span></span><span id="MJXc-Node-85" class="mjx-mi MJXc-space3"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.435em; padding-bottom: 0.291em; padding-right: 0.064em;">I</span></span><span id="MJXc-Node-86" class="mjx-mo"><span class="mjx-char MJXc-TeX-main-R" style="padding-top: 0.435em; padding-bottom: 0.579em;">(</span></span><span id="MJXc-Node-87" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.507em; padding-bottom: 0.291em;">h</span></span><span id="MJXc-Node-88" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.507em; padding-bottom: 0.291em;">k</span></span><span id="MJXc-Node-89" class="mjx-mi"><span class="mjx-char MJXc-TeX-math-I" style="padding-top: 0.507em; padding-bottom: 0.291em;">l</span></span><span id="MJXc-Node-90" class="mjx-mo"><span class="mjx-char MJXc-TeX-main-R" style="padding-top: 0.435em; padding-bottom: 0.579em;">)</span></span><span id="MJXc-Node-91" class="mjx-mo MJXc-space3"><span class="mjx-char MJXc-TeX-main-R" style="padding-top: 0.219em; padding-bottom: 0.363em;">></span></span></span></span></span></span></span><span class="MJX_Assistive_MathML" role="presentation"><math xmlns="http://www.w3.org/1998/Math/MathML" id="inf3"><mstyle displaystyle="true" scriptlevel="0"><mrow><mo><</mo><mi>I</mi><mo stretchy="false">(</mo><mi>h</mi><mi>k</mi><mi>l</mi><mo stretchy="false">)</mo><mo>></mo></mrow></mstyle></math></span></span><script type="math/mml" id="MathJax-Element-3"><math id="inf3"><mstyle displaystyle="true" scriptlevel="0"><mrow><mo><</mo><mi>I</mi><mo stretchy="false">(</mo><mi>h</mi><mi>k</mi><mi>l</mi><mo stretchy="false">)</mo><mo>></mo></mrow></mstyle></math></script> is the average intensity of that reflection.</p></div>
</li>
</ol>
</div>
Table Code Blocks
| data = {{},{},{}...,{}}; loglog=Function[{x,y},{Log[x/],Log[y]}]; datalog=Apply[loglog,data,{1}]; peakr = Function[{},Piecewise[{{+(1/2)*Log[1-(2/3)*] +*(1-(1-(2/3)*)^(3/2),<3/2/,Infinity}}]]; fit = NonlinearModelFit[datalog,{peakr[],{}}, {{},{},{}},x,Method->NMinimize]; fit['ParameterTable'] |
-
Table with Code Blocks
HTML
<div class="table">
<div class="table__table">
<table><tbody><tr><td align='left' valign='bottom'><span class='monospace'> data = </span><span class='monospace'>{{</span><math id='inf692'><mrow><msub><mi>x</mi><mn mathvariant='normal'>1</mn></msub><mo mathvariant='normal'>,</mo><msub><mi>R</mi><mn mathvariant='normal'>1</mn></msub></mrow></math><span class='monospace'>},{</span><math id='inf693'><mrow><msub><mi>x</mi><mn mathvariant='normal'>2</mn></msub><mo mathvariant='normal'>,</mo><msub><mi>R</mi><mn mathvariant='normal'>2</mn></msub></mrow></math><span class='monospace'>},{</span><math id='inf694'><mrow><msub><mi>x</mi><mn mathvariant='normal'>3</mn></msub><mo mathvariant='normal'>,</mo><msub><mi>R</mi><mn mathvariant='normal'>3</mn></msub></mrow></math><span class='monospace'>}...,{</span><math id='inf695'><mrow><msub><mi>x</mi><mi>m</mi></msub><mo mathvariant='normal'>,</mo><msub><mi>R</mi><mi>m</mi></msub></mrow></math><span class='monospace'>}};</span><br/><span class='monospace'>loglog=Function</span><span class='monospace'>[{x,y},{Log[x/</span><math id='inf6960'><msub><mrow><mo mathvariant='normal'stretchy='false'>[</mo><mrow><mi>C</mi><mo mathvariant='monospace'></mo><msup><mi>a</mi><mrow><mn mathvariant='normal'>2</mn><mo mathvariant='normal'>+</mo></mrow></msup></mrow><mo mathvariant='normal' stretchy='false'>]</mo></mrow><mn mathvariant='normal'>0</mn></msub></math><span class='monospace'>],Log[y]}];</span><br/><span class='monospace'>datalog=Apply[loglog,data,{1}];</span><br/><br/><br/><span class='monospace'>peakr = Function[{</span><math id='inf6970'><mrow><mi>A</mi><mo mathvariant='normal'>,</mo><mi>B</mi><mo mathvariant='normal'>,</mo><mi>C</mi><mo mathvariant='normal'>,</mo><mi>x</mi></mrow></math><span class='monospace'>},Piecewise[{{</span><math id='inf6980'><mi>A</mi></math><span class='monospace'>+(1/2)*Log[1-(2/3)*</span><math id='inf6990'><mrow><mi>B</mi><mo mathvariant='monospace'></mo><mi>x</mi></mrow></math><span class='monospace'>]</span><br/><span class='monospace'></span><span class='monospace'>+</span><math id='inf7000'><mi>C</mi></math><span class='monospace'>*(1-(1-(2/3)*</span><math id='inf7010'><mrow><mi>B</mi><mo mathvariant='monospace'></mo><mi>x</mi></mrow></math><span class='monospace'>)^(3/2),</span><math id='inf702'><mi mathvariant='normal'>x</mi></math><span class='monospace'><3/2/</span><math id='inf703'><mi>B</mi></math><span class='monospace'>,Infinity}}]];</span><br/><br/><br/><span class='monospace'>fit = NonlinearModelFit[datalog,{peakr[</span><math id='inf704'><mrow><mi>A</mi><mo mathvariant='normal'>,</mo><mi>B</mi><mo mathvariant='normal'>,</mo><mi>C</mi><mo mathvariant='normal'>,</mo><mi>x</mi></mrow></math><span class='monospace'>],{</span><math id='inf705'><mrow><mrow><mi>A</mi><mo mathvariant='normal'>></mo><mn mathvariant='normal'>0</mn></mrow><mo mathvariant='normal'>,</mo><mrow><mrow><mi>B</mi><mo mathvariant='normal'>></mo><mn mathvariant='normal'>0</mn></mrow><mo mathvariant='normal'>,</mo><mrow><mi>C</mi><mo mathvariant='normal'>></mo><mn mathvariant='normal'>0</mn></mrow></mrow></mrow></math><span class='monospace'>}},</span><br/><span class='monospace'></span><span class='monospace'>{{</span><math id='inf706'><mrow><mi>A</mi><mo mathvariant='normal'>,</mo><msub><mi>A</mi><mn mathvariant='normal'>0</mn></msub></mrow></math><span class='monospace'>},{</span><math id='inf707'><mrow><mi>B</mi><mo mathvariant='normal'>,</mo><msub><mi>B</mi><mn mathvariant='normal'>0</mn></msub></mrow></math><span class='monospace'>},{</span><math id='inf708'><mrow><mi>C</mi><mo mathvariant='normal'>,</mo><msub><mi>C</mi><mn mathvariant='normal'>0</mn></msub></mrow></math><span class='monospace'>}},x,Method->NMinimize];</span><br/><span class='monospace'>fit['ParameterTable']</span></td></tr></tbody></table>
</div>
<ol class="list">
<li class="table-footnote" >
<div class="table-footnote__text"><p>Table with Code Blocks</p></div>
</li>
</ol>
</div>
Table Equations
| Definitions of the simulated system | |
|---|---|
| Type | Status |
| Protein assembly occupancy | |
| Histones | |
| Regions of histones | |
-
Table with Equations
HTML
<div class="table">
<div class="table__table">
<table><thead><tr><th colspan='2' valign='top'>Definitions of the simulated system</th></tr><tr><th valign='top'>Type</th><th valign='top'>Status</th></tr></thead><tbody><tr><td valign='top'>Protein assembly occupancy<math id='inf11'><mstyle displaystyle='true' scriptlevel='0'><mrow><mi>k</mi><mo>∈</mo><mo stretchy='false'>[</mo><mn>1</mn><mo>,</mo><msub><mi>N</mi><mi>p</mi></msub><mo stretchy='false'>]</mo></mrow></mstyle></math></td><td valign='top'><math id='inf12'><mstyle displaystyle='true' scriptlevel='0'><mrow><msub><mi>P</mi><mrow><mi>k</mi></mrow></msub><mo>∈</mo><mrow><mo>{</mo><mrow><mi mathvariant='normal'>u</mi><mi mathvariant='normal'>n</mi><mi mathvariant='normal'>b</mi><mi mathvariant='normal'>o</mi><mi mathvariant='normal'>u</mi><mi mathvariant='normal'>n</mi><mi mathvariant='normal'>d</mi><mo>,</mo><mspace width='thinmathspace'/><mi mathvariant='normal'>b</mi><mi mathvariant='normal'>o</mi><mi mathvariant='normal'>u</mi><mi mathvariant='normal'>n</mi><mi mathvariant='normal'>d</mi></mrow><mo>}</mo></mrow></mrow></mstyle></math></td></tr><tr><td valign='top'>Histones<math id='inf13'><mstyle displaystyle='true'scriptlevel='0'><mrow><mi>i</mi><mo>∈</mo><mo stretchy='false'>[</mo><mn>1</mn><mo>,</mo><mi>N</mi><mo stretchy='false'>]</mo></mrow></mstyle></math></td><td valign='top'><math id='inf14'><mstyle displaystyle='true' scriptlevel='0'><mrow><msub><mi>S</mi><mrow><mi>i</mi></mrow></msub><mo>∈</mo><mrow><mo>{</mo><mrow><mi mathvariant='normal'>m</mi><mi mathvariant='normal'>e</mi><mn>0</mn><mo>,</mo><mspace width='thinmathspace'/><mi mathvariant='normal'>m</mi><mi mathvariant='normal'>e</mi><mn>1</mn><mo>,</mo><mspace width='thinmathspace'/><mi mathvariant='normal'>m</mi><mi mathvariant='normal'>e</mi><mn>2</mn><mo>,</mo><mspace width='thinmathspace'/><mi mathvariant='normal'>m</mi><mi mathvariant='normal'>e</mi><mn>3</mn></mrow><mo>}</mo></mrow></mrow></mstyle></math></td></tr><tr><td valign='top'>Regions of histones</td><td><math id='inf15'><mstyle displaystyle='true' scriptlevel='0'><mrow><msub><mi>R</mi><mrow><mi>m</mi></mrow></msub><mo>=</mo><mrow><mo>{</mo><mtable columnspacing='1em' rowspacing='4pt'><mtr><mtd><mrow><mi mathvariant='normal'>H</mi><mi mathvariant='normal'>i</mi><mi mathvariant='normal'>s</mi><mi mathvariant='normal'>t</mi><mi mathvariant='normal'>o</mi><mi mathvariant='normal'>n</mi><mi mathvariant='normal'>e</mi><mi mathvariant='normal'>s</mi></mrow><mo stretchy='false'>[</mo><mn>1</mn><mo>,</mo><mo>.</mo><mo>.</mo><mo>.</mo><mo>,</mo><msub><mi>N</mi><mrow><mi>N</mi><mi>R</mi></mrow></msub><mo stretchy='false'>]</mo><mspace width='thinmathspace'/><mrow><mi mathvariant='normal'>w</mi><mi mathvariant='normal'>h</mi><mi mathvariant='normal'>e</mi><mi mathvariant='normal'>n</mi></mrow><mspace width='thinmathspace'/><mi>m</mi><mo>=</mo><mi>N</mi><mi>R</mi><mspace width='thinmathspace'/><mrow><mo stretchy='false'>(</mo><mi mathvariant='normal'>n</mi><mi mathvariant='normal'>u</mi><mi mathvariant='normal'>c</mi><mi mathvariant='normal'>l</mi><mi mathvariant='normal'>e</mi><mi mathvariant='normal'>a</mi><mi mathvariant='normal'>t</mi><mi mathvariant='normal'>i</mi><mi mathvariant='normal'>o</mi><mi mathvariant='normal'>n</mi><mspace width='thinmathspace'/><mi mathvariant='normal'>r</mi><mi mathvariant='normal'>e</mi><mi mathvariant='normal'>g</mi><mi mathvariant='normal'>i</mi><mi mathvariant='normal'>o</mi><mi mathvariant='normal'>n</mi><mo stretchy='false'>)</mo></mrow></mtd></mtr><mtr><mtd><mrow><mi mathvariant='normal'>H</mi><mi mathvariant='normal'>i</mi><mi mathvariant='normal'>s</mi><mi mathvariant='normal'>t</mi><mi mathvariant='normal'>o</mi><mi mathvariant='normal'>n</mi><mi mathvariant='normal'>e</mi><mi mathvariant='normal'>s</mi></mrow><mo stretchy='false'>[</mo><msub><mi>N</mi><mrow><mi>N</mi><mi>R</mi></mrow></msub><mo>+</mo><mn>1</mn><mo>,</mo><mo>.</mo><mo>.</mo><mo>.</mo><mo>,</mo><mi>N</mi><mo stretchy='false'>]</mo><mspace width='thinmathspace'/><mrow><mi mathvariant='normal'>w</mi><mi mathvariant='normal'>h</mi><mi mathvariant='normal'>e</mi><mi mathvariant='normal'>n</mi></mrow><mspace width='thinmathspace'/><mi>m</mi><mo>=</mo><mi>B</mi><mspace width='thinmathspace'/><mrow><mo stretchy='false'>(</mo><mi mathvariant='normal'>b</mi><mi mathvariant='normal'>o</mi><mi mathvariant='normal'>d</mi><mi mathvariant='normal'>y</mi><mspace width='thinmathspace'/><mi mathvariant='normal'>r</mi><mi mathvariant='normal'>e</mi><mi mathvariant='normal'>g</mi><mi mathvariant='normal'>i</mi><mi mathvariant='normal'>o</mi><mi mathvariant='normal'>n</mi><mo stretchy='false'>)</mo></mrow><mo>,</mo><mspace width='thinmathspace'/><msub><mi>N</mi><mi>B</mi></msub><mo>=</mo><mi>N</mi><mo>−</mo><msub><mi>N</mi><mrow><mi>N</mi><mi>R</mi></mrow></msub></mtd></mtr><mtr><mtd><mrow><mrow><mi mathvariant='normal'>H</mi><mi mathvariant='normal'>i</mi><mi mathvariant='normal'>s</mi><mi mathvariant='normal'>t</mi><mi mathvariant='normal'>o</mi><mi mathvariant='normal'>n</mi><mi mathvariant='normal'>e</mi><mi mathvariant='normal'>s</mi></mrow><mo stretchy='false'>[</mo><mn>1</mn><mo>,</mo><mo>.</mo><mo>.</mo><mo>.</mo><mo>,</mo><mi>N</mi><mo stretchy='false'>]</mo><mspace width='thinmathspace'/><mrow><mi mathvariant='normal'>w</mi><mi mathvariant='normal'>h</mi><mi mathvariant='normal'>e</mi><mi mathvariant='normal'>n</mi></mrow><mspace width='thinmathspace'/><mi>m</mi><mo>=</mo><mi>L</mi><mspace width='thinmathspace'/><mo stretchy='false'>(</mo><mi mathvariant='normal'>e</mi><mi mathvariant='normal'>n</mi><mi mathvariant='normal'>t</mi><mi mathvariant='normal'>i</mi><mi mathvariant='normal'>r</mi><mi mathvariant='normal'>e</mi><mspace width='thinmathspace'/><mi mathvariant='normal'>r</mi><mi mathvariant='normal'>e</mi><mi mathvariant='normal'>g</mi><mi mathvariant='normal'>i</mi><mi mathvariant='normal'>o</mi><mi mathvariant='normal'>n</mi><mo stretchy='false'>)</mo></mrow><mo>,</mo><mspace width='thinmathspace'/><msub><mi>N</mi><mi>L</mi></msub><mo>=</mo><mi>N</mi></mtd></mtr></mtable><mo fence='true' stretchy='true' symmetric='true'/></mrow></mrow></mstyle></math></td></tr></tbody></table>
</div>
<ol class="list">
<li class="table-footnote" >
<div class="table-footnote__text"><p>Table with Equations</p></div>
</li>
</ol>
</div>
Table Gene Sequences
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Recombinant DNA reagent | synthesized gene - mScarletI | Genscript | actagtggtggttcaggaGTTTCAAAAGGTGAAGCCGTTATTAAAGAATTTATGAGATTCAAGGTTCACATGGAAGGAAGTATGAACGGTCATGAATTTGAGATTGAAGGAGAAGGTGAAGGTAGACCATATGAAGGCACCCAAACAGCTAAATTAAAAGTAACTAAAGGTGGTCCATTACCATTTAGTTGGGATATTTTATCTCCACAATTTATGTATGGTTCACGTGCTTTCAttAAACATCCAGCAGATATTCCAGATTATTATAAACAATCATTTCCAGAAGGTTTTAAATGGGAACGTGTCATGAACTTTGAAGATGGTGGAGCAGTTACAGTCACACAAGATACCTCATTAGAAGATGGTACATTAATATATAAAGTTAAATTACGTGGTACTAATTTTCCACCAGACGGTCCAGTAATGCAAAAAAAAACAATGGGCTGGGAAGCTAGTACAGAACGTTTATATCCTGAAGATGGTGTCCTTAAAGGCGATATAAAAATGGCCTTGAGATTAAAGGATGGTGGTAGGTATTTAGCAGATTTCAAAACCACTTATAAAGCAAAAAAACCAGTTCAAATGCCAGGTGCATATAATGTTGATAGAAAACTTGATATTACCAGTCATAATGAAGATTACACAGTTGTCGAACAATACGAACGTTCTGAAGGTCGTCATAGCACTGGTGGTATGGATGAATTATACAAATAAgctagc | d |
| Recombinant DNA reagent | synthesized gene - mNeon | Genscript | actagtggtggttcaggaGTTTCAAAAGGTGAAGCCGTTATTAAAGAATTTATGAGATTCAAGGTTCACATGGAAGGAAGTATGAACGGTCATGAATTTGAGATTGAAGGAGAAGGTGAAGGTAGACCATATGAAGGCACCCAAACAGCTAAATTAAAAGTAACTAAAGGTGGTCCATTACCATTTAGTTGGGATATTTTATCTCCACAATTTATGTATGGTTCACGTGCTTTCAttAAACATCCAGCAGATATTCCAGATTATTATAAACAATCATTTCCAGAAGGTTTTAAATGGGAACGTGTCATGAACTTTGAAGATGGTGGAGCAGTTACAGTCACACAAGATACCTCATTAGAAGATGGTACATTAATATATAAAGTTAAATTACGTGGTACTAATTTTCCACCAGACGGTCCAGTAATGCAAAAAAAAACAATGGGCTGGGAAGCTAGTACAGAACGTTTATATCCTGAAGATGGTGTCCTTAAAGGCGATATAAAAATGGCCTTGAGATTAAAGGATGGTGGTAGGTATTTAGCAGATTTCAAAACCACTTATAAAGCAAAAAAACCAGTTCAAATGCCAGGTGCATATAATGTTGATAGAAAACTTGATATTACCAGTCATAATGAAGATTACACAGTTGTCGAACAATACGAACGTTCTGAAGGTCGTCATAGCACTGGTGGTATGGATGAATTATACAAATAAgctagc |
-
Table with Gene Sequences
HTML
<div class="table">
<div class="table__table">
<table><thead><tr><th>Reagent type (species) or resource</th><th>Designation</th><th>Source or reference</th><th>Identifiers</th><th>Additional information</th></tr></thead><tbody><tr><td>Recombinant DNA reagent</td><td>synthesized gene - mScarletI</td><td>Genscript</td><td><span class='sequence'>actagtggtggttcaggaGTTTCAAAAGGTGAAGCCGTTATTAAAGAATTTATGAGATTCAAGGTTCACATGGAAGGAAGTATGAACGGTCATGAATTTGAGATTGAAGGAGAAGGTGAAGGTAGACCATATGAAGGCACCCAAACAGCTAAATTAAAAGTAACTAAAGGTGGTCCATTACCATTTAGTTGGGATATTTTATCTCCACAATTTATGTATGGTTCACGTGCTTTCAttAAACATCCAGCAGATATTCCAGATTATTATAAACAATCATTTCCAGAAGGTTTTAAATGGGAACGTGTCATGAACTTTGAAGATGGTGGAGCAGTTACAGTCACACAAGATACCTCATTAGAAGATGGTACATTAATATATAAAGTTAAATTACGTGGTACTAATTTTCCACCAGACGGTCCAGTAATGCAAAAAAAAACAATGGGCTGGGAAGCTAGTACAGAACGTTTATATCCTGAAGATGGTGTCCTTAAAGGCGATATAAAAATGGCCTTGAGATTAAAGGATGGTGGTAGGTATTTAGCAGATTTCAAAACCACTTATAAAGCAAAAAAACCAGTTCAAATGCCAGGTGCATATAATGTTGATAGAAAACTTGATATTACCAGTCATAATGAAGATTACACAGTTGTCGAACAATACGAACGTTCTGAAGGTCGTCATAGCACTGGTGGTATGGATGAATTATACAAATAAgctagc</span></td><td>d</td></tr><tr><td>Recombinant DNA reagent</td><td>synthesized gene - mNeon</td><td>Genscript</td><td><span class='sequence'>actagtggtggttcaggaGTTTCAAAAGGTGAAGCCGTTATTAAAGAATTTATGAGATTCAAGGTTCACATGGAAGGAAGTATGAACGGTCATGAATTTGAGATTGAAGGAGAAGGTGAAGGTAGACCATATGAAGGCACCCAAACAGCTAAATTAAAAGTAACTAAAGGTGGTCCATTACCATTTAGTTGGGATATTTTATCTCCACAATTTATGTATGGTTCACGTGCTTTCAttAAACATCCAGCAGATATTCCAGATTATTATAAACAATCATTTCCAGAAGGTTTTAAATGGGAACGTGTCATGAACTTTGAAGATGGTGGAGCAGTTACAGTCACACAAGATACCTCATTAGAAGATGGTACATTAATATATAAAGTTAAATTACGTGGTACTAATTTTCCACCAGACGGTCCAGTAATGCAAAAAAAAACAATGGGCTGGGAAGCTAGTACAGAACGTTTATATCCTGAAGATGGTGTCCTTAAAGGCGATATAAAAATGGCCTTGAGATTAAAGGATGGTGGTAGGTATTTAGCAGATTTCAAAACCACTTATAAAGCAAAAAAACCAGTTCAAATGCCAGGTGCATATAATGTTGATAGAAAACTTGATATTACCAGTCATAATGAAGATTACACAGTTGTCGAACAATACGAACGTTCTGAAGGTCGTCATAGCACTGGTGGTATGGATGAATTATACAAATAAgctagc</span></td><td></td></tr></tbody></table>
</div>
<ol class="list">
<li class="table-footnote" >
<div class="table-footnote__text"><p>Table with Gene Sequences</p></div>
</li>
</ol>
</div>
Table Inline Images
| unwinding | turnover | ||
|---|---|---|---|
| # | Chemical drawing | IC50[95%CI];µM | IC50[95%CI];µM |
| 2 | 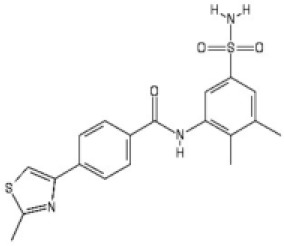 | 2.2 [1.7–2.7] | 3.2 [2.3–4.0] |
-
Table with inline images
HTML
<div class="table">
<div class="table__table">
<table><thead><tr><th/><th/><th>unwinding</th><th>turnover</th></tr></thead><tbody><tr><td>#</td><td>Chemical drawing</td><td>IC<sub>50</sub>[95%CI];µM</td><td>IC<sub>50</sub>[95%CI];µM</td></tr><tr><td>2</td><td><img src='https://cdn.elifesciences.org/articles/65339/elife-65339-inf1-v1.jpg'/></td><td>2.2<br/>[1.7–2.7]</td><td>3.2<br/>[2.3–4.0]</td></tr></tbody></table>
</div>
<ol class="list">
<li class="table-footnote" >
<div class="table-footnote__text"><p>Table with inline images</p> </div>
</li>
</ol>
</div>
Table Long Url
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Software, algorithm | Metamorph | Metamorph v | https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy#gref |
-
Table with long url
HTML
<div class="table">
<div class="table__table">
<table><thead><tr><th align='left' valign='bottom'>Reagent type (species) or resource</th><th align='left' valign='bottom'>Designation</th><th align='left' valign='bottom'>Source or reference</th><th align='left' valign='bottom'>Identifiers</th><th align='left' valign='bottom'>Additional information</th></tr></thead><tbody><tr><td align='left' valign='bottom'>Software, algorithm</td><td align='left' valign='bottom'>Metamorph</td><td align='left' valign='bottom'>Metamorph v</td><td align='left' valign='bottom'><a href='https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy#gref'>https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy#gref</a></td><td></td></tr></tbody></table>
</div>
<ol class="list">
<li class="table-footnote" >
<div class="table-footnote__text"><p>Table with long url</p></div>
</li>
</ol>
</div>
Table Ordered List
| Factors classically associated with immune functions | ||||
|---|---|---|---|---|
| Protein | Immune system properties | Nervous system properties | References | |
| Antimicrobial peptides(AMPs) |
|
| Hanson et al., 2019;Lezi et al., 2018; | |
-
Table with ordered list
HTML
<div class="table">
<div class="table__table">
<table><thead><tr><th colspan='5' valign='top'>Factors classically associated with immune functions</th></tr><tr valign='bottom'><th colspan='2'>Protein</th><th>Immune system properties</th><th>Nervous system properties</th><th>References</th></tr></thead><tbody><tr valign='top'><td colspan='2'>Antimicrobial peptides(AMPs)</td><td><ol class='listlist--number'><li><p>Secreted by epithelial and phagocytic cells</p></li><li><p>Disrupt microbial membranes leading to destruction of pathogen</p></li></ol></td><td><ol class='listlist--number'><li><p>Antimicrobial in nervous system niches</p></li><li><p>Control chemotaxis of immune cells and astroglia</p></li><li><p>Mediate iron homeostasis</p></li><li><p>Modulate nerve impulses</p></li><li><p>Implicated in aging and neurodegeneration</p></li></ol></td><td><a href='#bib107'>Hanson et al., 2019</a>;<a href='#bib161'>Lezi et al., 2018</a>;<ahref='#bib246'>Su et al., 2010</a>;<ahref='#bib290'>Zasloff, 2002</a></td></tr></tbody></table>
</div>
<ol class="list">
<li class="table-footnote" >
<div class="table-footnote__text"><p>Table with ordered list</p></div>
</li>
</ol>
</div>
Table Unordered List
| Factors classically associated with immune functions | ||||
|---|---|---|---|---|
| Protein | Immune system properties | Nervous system properties | References | |
| Antimicrobial peptides(AMPs) |
|
| Hanson et al., 2019;Lezi et al., 2018;Su et al., 2010;Zasloff, 2002 | |
-
Table with unordered list
HTML
<div class="table">
<div class="table__table">
<table><thead><tr><th colspan='5' valign='top'>Factors classically associated with immune functions</th></tr><tr valign='bottom'><th colspan='2'>Protein</th><th>Immune system properties</th><th>Nervous system properties</th><th>References</th></tr></thead><tbody><tr valign='top'><td colspan='2'>Antimicrobial peptides(AMPs)</td><td><ul class='listlist--bullet'><li><p>Secreted by epithelial and phagocyticcells</p></li><li><p>Disrupt microbial membranes leading to destruction of pathogen</p></li></ul></td><td><ul class='listlist--bullet'><li><p>Antimicrobial in nervous system niches</p></li><li><p>Control chemotaxis of immune cells and astroglia</p></li><li><p>Mediate iron homeostasis</p></li><li><p>Modulate nerve impulses</p></li><li><p>Implicated in aging and neurodegeneration</p></li></ul></td><td><a href='#bib107'>Hanson et al., 2019</a>;<a href='#bib161'>Lezi et al., 2018</a>;<a href='#bib246'>Su et al., 2010</a>;<a href='#bib290'>Zasloff, 2002</a></td></tr></tbody></table>
</div>
<ol class="list">
<li class="table-footnote" >
<div class="table-footnote__text"><p>Table with unordered list</p></div>
</li>
</ol>
</div>
Term
HTML
<span class="term">Important</span>
Term Highlighted
HTML
<span class="term term-highlighted">Important</span>
To Top Link
HTML
<a href="#siteHeader" class="to-top-link">Back to top</a>
Tweet
atoms-date
HTML
<article class="tweet" data-behaviour="Twitter">
<blockquote class="tweet__content twitter-tweet">
<p class="paragraph">Tune in to the latest <a href="https://twitter.com/hashtag/eLifePodcast?src=hash&ref_src=twsrc%5Etfw">#eLifePodcast</a> to hear authors discuss the stories behind their recent discoveries <a href="https://t.co/awwqu5L3lE">https://t.co/awwqu5L3lE</a></p>
<div class="tweet__by" aria-label="Tweeted by">eLife - the journal (@eLife)</div>
<div class="tweet__context">
<span class="date"> <time datetime="2020-02-07">Feb 7, 2020</time></span>
<a class="tweet__link" href="https://twitter.com/eLife/status/1225868108168093696">See tweet</a>
</div>
</blockquote>
</article>
Tweet With Cards
Tune in to the latest #eLifePodcast to hear authors discuss the stories behind their recent discoveries https://t.co/awwqu5L3lE
eLife - the journal (@eLife)
atoms-date
HTML
<article class="tweet" data-behaviour="Twitter">
<blockquote class="tweet__content twitter-tweet">
<p class="paragraph">Tune in to the latest <a href="https://twitter.com/hashtag/eLifePodcast?src=hash&ref_src=twsrc%5Etfw">#eLifePodcast</a> to hear authors discuss the stories behind their recent discoveries <a href="https://t.co/awwqu5L3lE">https://t.co/awwqu5L3lE</a></p>
<div class="tweet__by" aria-label="Tweeted by">eLife - the journal (@eLife)</div>
<div class="tweet__context">
<span class="date"> <time datetime="2020-02-07">Feb 7, 2020</time></span>
<a class="tweet__link" href="https://twitter.com/eLife/status/1225868108168093696">See tweet</a>
</div>
</blockquote>
</article>
Tweet With Conversation
I love it when people show interest in #EvoMed!
Bridget S. Preston, Esq. (@bridgetmcgann)
atoms-date
HTML
<article class="tweet" data-behaviour="Twitter">
<blockquote class="tweet__content twitter-tweet">
<p class="paragraph">I love it when people show interest in <a href="https://twitter.com/hashtag/EvoMed?src=hash&ref_src=twsrc%5Etfw">#EvoMed</a>!</p>
<div class="tweet__by" aria-label="Tweeted by">Bridget S. Preston, Esq. (@bridgetmcgann)</div>
<div class="tweet__context">
<span class="date"> <time datetime="2020-02-13">Feb 13, 2020</time></span>
<a class="tweet__link" href="https://twitter.com/bridgetmcgann/status/1227803569547612162">See tweet</a>
</div>
</blockquote>
</article>
Video
HTML
<video controls="controls" poster="https://static-movie-usa.glencoesoftware.com/jpg/10.7554/872/946920eb226e045af1781fe292f2cd5206b30a75/elife-14093-app2-media1.jpg" preload="metadata">
<img src="https://static-movie-usa.glencoesoftware.com/jpg/10.7554/872/946920eb226e045af1781fe292f2cd5206b30a75/elife-14093-app2-media1.jpg" alt="posterframe for video" />
<p>This video cannot be played in place because your browser does support HTML5 video. You may still download the video for offline viewing.</p>
<source src="https://static-movie-usa.glencoesoftware.com/mp4/10.7554/872/946920eb226e045af1781fe292f2cd5206b30a75/elife-14093-app2-media1.mp4" type='video/mp4; codecs="avc1.42E01E, mp4a.40.2"' class="media-source"/>
<div>
<a class="media-source__fallback_link" href="https://static-movie-usa.glencoesoftware.com/mp4/10.7554/872/946920eb226e045af1781fe292f2cd5206b30a75/elife-14093-app2-media1.mp4">Download as mp4</a>
</div>
<source src="https://static-movie-usa.glencoesoftware.com/webm/10.7554/872/946920eb226e045af1781fe292f2cd5206b30a75/elife-14093-app2-media1.webm" type='video/webm; codecs="vp8.0, vorbis"' class="media-source"/>
<div>
<a class="media-source__fallback_link" href="https://static-movie-usa.glencoesoftware.com/webm/10.7554/872/946920eb226e045af1781fe292f2cd5206b30a75/elife-14093-app2-media1.webm">Download as webm</a>
</div>
<source src="https://static-movie-usa.glencoesoftware.com/ogv/10.7554/872/946920eb226e045af1781fe292f2cd5206b30a75/elife-14093-app2-media1.ogv" type='video/ogg; codecs="theora, vorbis"' class="media-source"/>
<div>
<a class="media-source__fallback_link" href="https://static-movie-usa.glencoesoftware.com/ogv/10.7554/872/946920eb226e045af1781fe292f2cd5206b30a75/elife-14093-app2-media1.ogv">Download as ogg</a>
</div>
</video>

